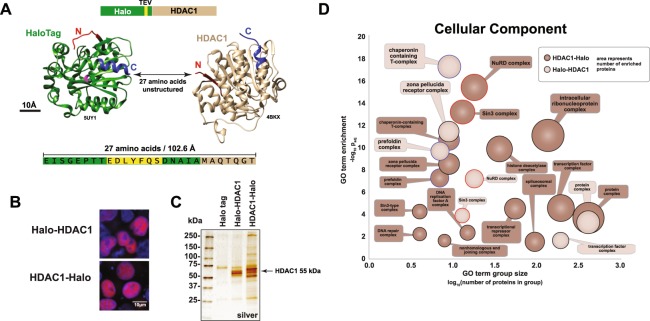
Figure 1.
Protein complexes associated with Halo-tagged HDAC1 expressed in HeLa cells. (A) Both the N and C termini of HDAC1 are potentially available for affinity tagging. The structures of HaloTag (green, ligand binding site in pink, PDB ID 5UY160) and human HDAC1 (brown, PDB ID 4BKX61) visualized using Chimera62 are shown to scale. The 27 amino-acid region in Halo-HDAC1 not present in the structures is also shown to scale. An additional 106 amino acids at the C terminus of HDAC1 are absent from the structure. (B) Localization of Halo-HDAC1 and HDAC1-Halo in HEK293T cells. Halo-tagged proteins are labeled with TMRDirect™ ligand (red). Nuclei are stained with Hoechst dye (blue). (C) Proteins enriched by either Halo-HDAC1 or HDAC1-Halo in HeLa cells. Proteins were resolved on a 4–15% SDS-PAGE gel and visualized by silver staining. Similar samples were analysed by Western blotting using HDAC1 antibodies to confirm the presence of full length HDAC1 in the eluates (Supplementary Figure 1). (D) Protein complexes copurifying with Halo-HDAC1 or HDAC1-Halo. Proteins significantly enriched (log2FC > 2, FDRup < 0.05) in Halo-HDAC1 or HDAC1-Halo purifications (3 biological replicates each) compared with controls expressing the Halo tag alone (3 biological replicates) were analysed for enriched GO terms (cellular component) using DAVID32 (Supplementary Table S2). Significantly enriched GO terms (padj < 0.05) which include the word “complex” are shown.