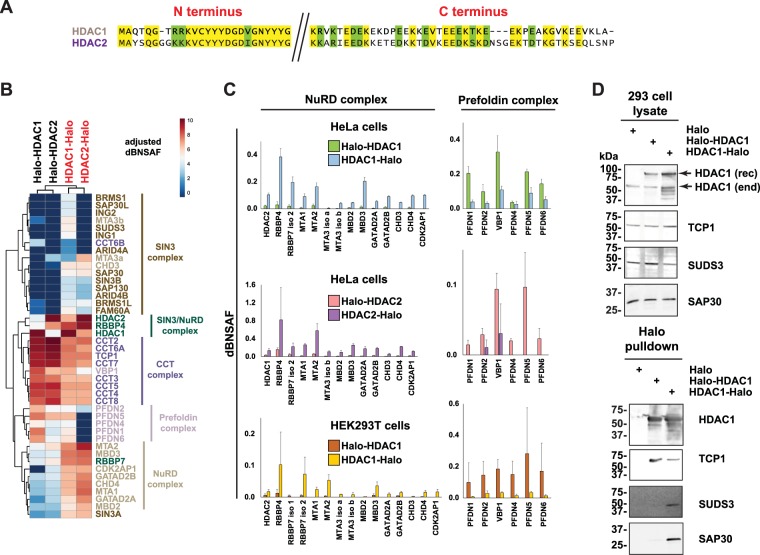
Figure 3.
HDAC2 (HeLa) or HDAC1 (HEK293T) associations with Sin3, NuRD, CCT and prefoldin complexes depends on affinity tag location. (A) Sequence alignment of the N and C termini of human HDAC1 (NP_004955) and human HDAC2 (NP_001518), using the AlignX tool in Vector NTI64. Identical residues are highlighted in yellow and similar residues in green. (B) Hierarchical clustering of Halo-tagged HDAC1 and HDAC2 associated complexes. Bait normalized dNSAF values (Supplementary Tables S3 and S4) were scaled 1000x prior to clustering using R. Values were further adjusted using a log2 transformation for representation using the indicated colour scale. Dissimilarity matrix calculations were based on Euclidean distance. (C) Components of the NuRD or prefoldin complexes copurifying with tagged versions of either HDAC1 (HeLa cells), HDAC2 (HeLa cells) or with HDAC1 (HEK293T cells). Error bars indicate standard deviation. (D) Lysates from HEK293T cells transfected with constructs expressing either Halo tag alone, Halo-HDAC1 or HDAC1-Halo were processed as described in “Methods”. Halo purified samples were analysed using SDS-PAGE and visualised by Western blotting using antibodies to the proteins indicated. Bands corresponding to recombinant Halo-tagged HDAC1 (HDAC1(rec)) or endogenous HDAC1 (HDAC1(end)) in the lysate samples are marked. Full length images of Western blots are presented in Supplementary Figure 2.