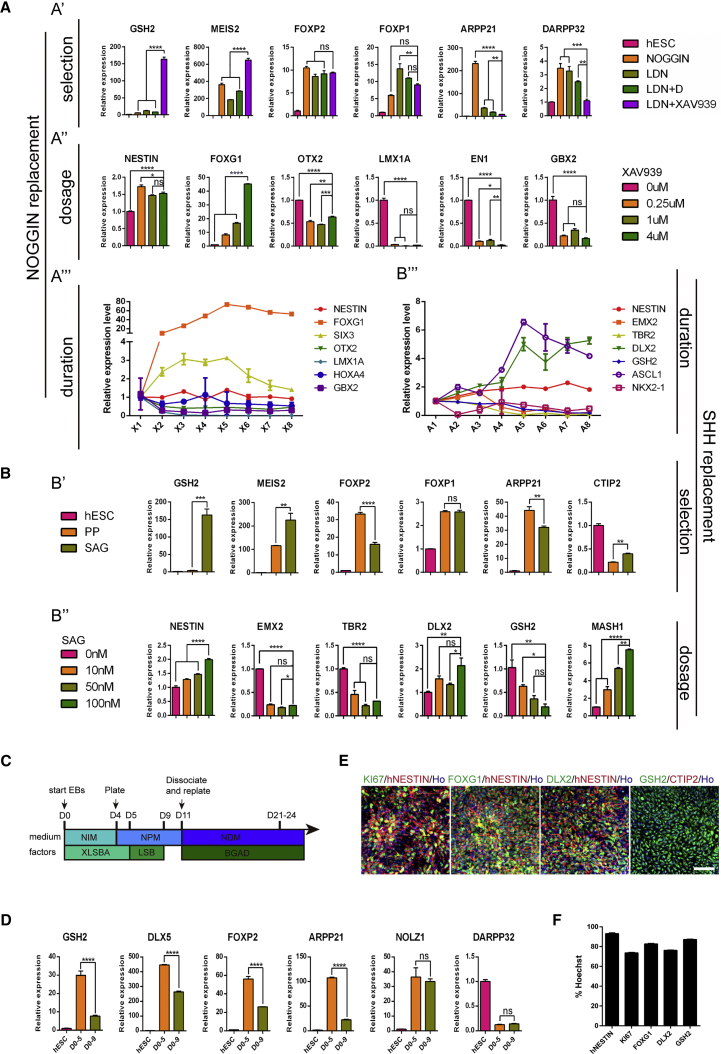
Figure 2.
Generation of LGE-like Progenitors from hESCs by Chemical Cocktails
(A) Relative mRNA expression of striatal lineage genes to identify optimal small-molecule combination to replace noggin. (A′) Striatal lineage gene expression in H9-hESCs cultures differentiated in the presence of LDN-193193 (100 nM), dorsomorphin (100 nM), XAV939 (1 μM), or their combination from days 1–9. All expression levels are normalized to levels detected in hESCs. (A″) Anterior-posterior axis gene expression in H9-hESC cultures differentiated after treatment with different concentrations of XAV939 (0, 0.25, 1, and 4 μM). (A‴) Anterior-posterior axis gene expression in H9-hESCs cultures differentiated after treatment with 4 μM XAV939 for different time periods. X1 represents cell cultures treated with XAV939 for 1 day, X2 represents cell cultures treated for 2 days, and so on. Data are presented as mean ± SEM (n = 3). ns, not significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; one-way ANOVA, followed by Tukey's multiple comparisons tests.
(B) Relative mRNA expression of striatal lineage genes to identify optimal small molecule to replace SHH. (B′) Striatal lineage gene expression in H9-hESC cultures differentiated in the presence of purmorphamine (PP, 1 μM) or SAG (100 nM). All expression levels are normalized to levels detected in hESCs. (B″) Dorsal-ventral axis gene expression in H9-hESCs cultures differentiated after treatment with different concentrations of SAG (0, 10, 50, and 100 nM). (B‴) Dorsal-ventral axis gene expression in H9-hESCs cultures differentiated after treatment with 100 nM SAG for different time periods. A1 represents cell cultures treated with SAG for 1 day. A2 represents cell cultures treated for 2 days, and so on. Data are presented as mean ± SEM (n = 3). ns, not significant. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; one-way ANOVA followed by Tukey's multiple comparisons tests.
(C) Schematic overview of the optimized differentiation protocol for hESCs.
(D) Relative mRNA expression of striatal lineage genes treated with small molecules from days 0–5 and days 0–9. All expression levels are normalized to levels detected in hESCs. Data are presented as mean ± SEM. ns, no significant. ∗∗∗∗p < 0.0001; Student's t test.
(E) Immunostaining images for neural progenitor marker NESTIN, proliferative marker KI67, forebrain marker FOXG1, LGE progenitor markers GSH2 and DLX2, and post-mitotic MSN marker CTIP2 on day 11. Scale bar, 50 μm.
(F) Quantification for immunostaining of markers in (E). Error bar represents SEM.