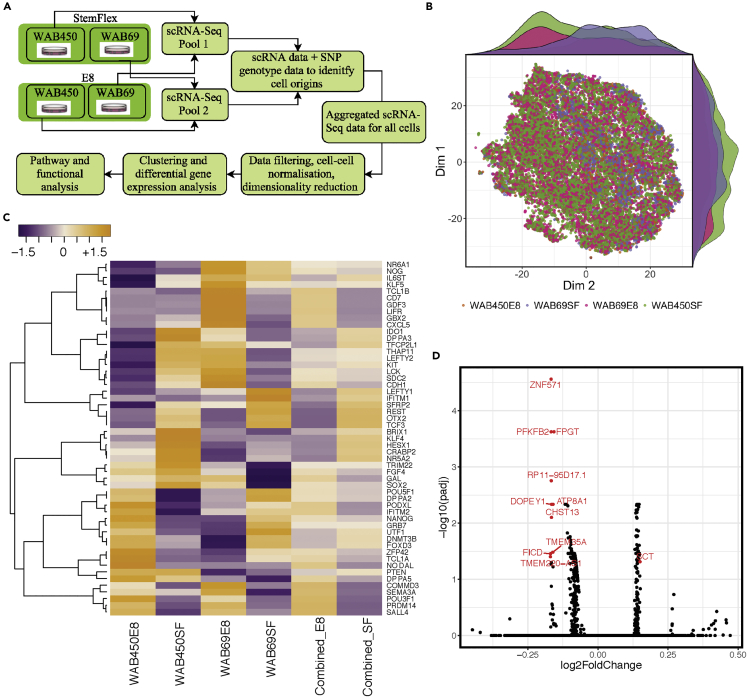
Figure 3.
Comparing Effects of the Two Media at Single-Cell Level
(A) Schematic representation of the experimental randomization design. Two cell lines were cultured separately in two media. The four cell cultures were genotyped (HumanCore Beadchip arrays) and were randomly combined into two pools for scRNA-seq experiment (10X Chromium). Cells were assigned to original sample based on sample SNP genotype and single-cell SNPs called from scRNA-seq data. The scRNA-seq data were aggregated for all four samples, filtered, normalized, clustered, and analyzed for differential gene expression and functional pathways.
(B) Two-dimensional distribution of cells based on gene expression profile. Dimensions 1 and 2 are principal coordinates of imputed values from CIDR (Clustering through Imputation and Dimensionality Reduction) (Lin et al., 2017). The overlap between the four culturing samples suggests the overall similar effects of the two media.
(C) Heatmap analysis of known pluripotency markers shown for four separate samples or the combination of two samples. Standardized gene expression is shown from low (purple color) to high (orange color). The numbers of genes expressed highly in StemFlex or in TeSR-E8 are similar, suggesting the two media maintain similar pluripotency states.
(D) Volcano plot shows genome-wide differential expression results between cells in StemFlex versus cells in TeSR-E8 (positive log2Folchange indicates higher expression in StemFlex). The numbers of upregulated genes in each media are similar. Red text indicates genes for which upregulation is statistically significant between the two conditions.