Figure 1.
A Meta-Analysis of TFs Enrichment in dNSCs Highlights Their Association with Distinct Neural Lineages
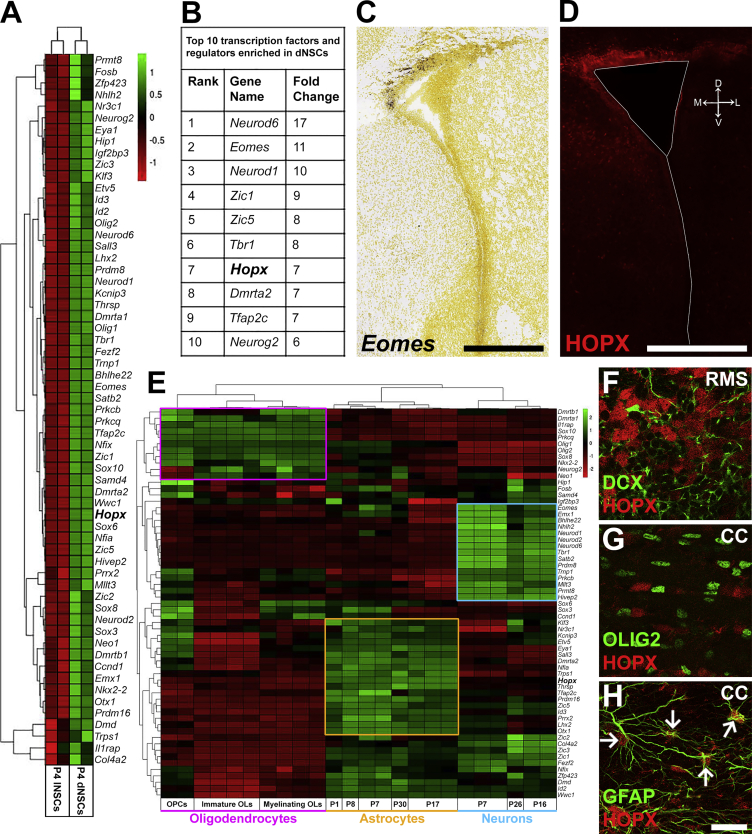
(A) Heatmap showing enrichment of 61 TFs in dNSCs compared with lNSCs (≥1.8-fold and p < 0.05).
(B) Top ten TFs enriched in dNSCs.
(C and D): Dorsal enrichment of select transcripts was confirmed using the Allen Brain Atlas for Eomes (C) and by immunohistochemistry for HOPX (D).
(E) Heatmap of dNSC enriched TFs reveals three clusters corresponding to defined neural lineages: oligodendrocytes (purple, 11/61); astrocytes (yellow, 18/61); neurons (turquoise, 15/61). Hopx (highlighted in bold) associates with the astrocytic lineage.
(F–H) Confirmation of astroglial lineage-specific enrichment of HOPX by immunohistochemistry. HOPX is largely absent in neuroblasts of the RMS (DCX; F) and oligodendrocytes in the CC (OLIG2; G), but is observed in astrocytes of the CC (GFAP; H, arrows indicate double positive cells).
CC, corpus callosum; dNSC, dorsal NSCs; lNSC, lateral NSCs; RMS, rostral migratory stream; OPC, oligodendrocyte precursor cell; OL, oligodendrocyte. Scale bars, 500 μm (C and D) and 25 μm (H).