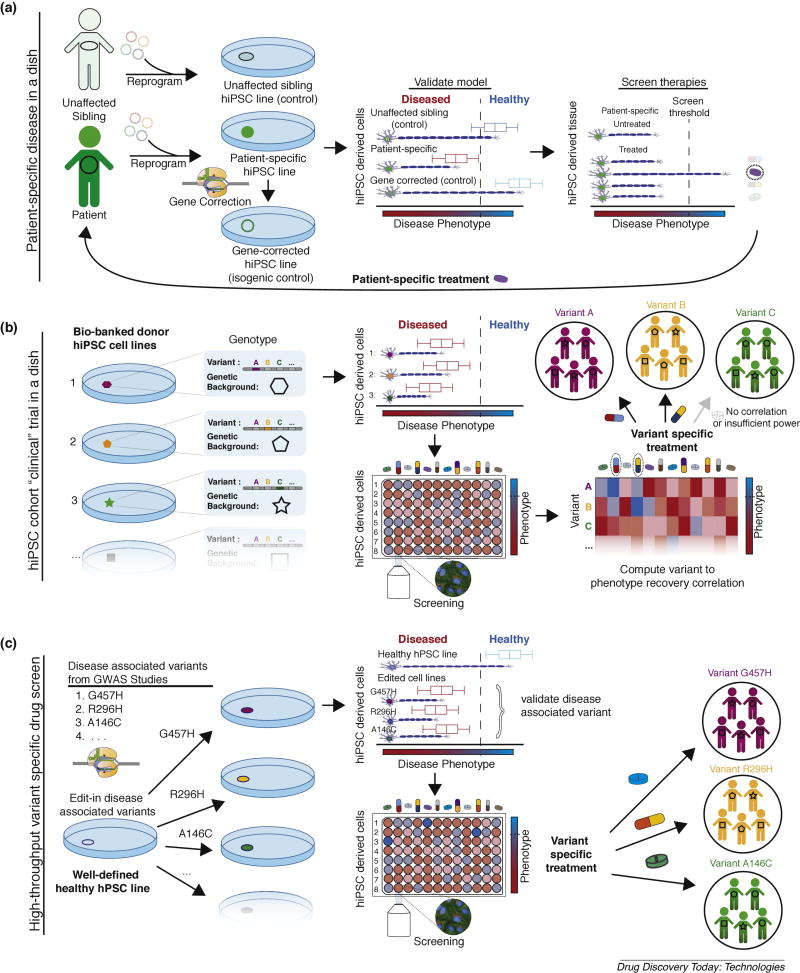
Figure 1. Paradigms of stem cell disease modeling for therapeutic screening.
(A) Patient-specific disease in a dish models. Unaffected siblings share only ~50% genetic inheritance with affected patients and the field has largely moved towards isogenic controls to validate phenotypic outputs (diseased vs. healthy). These screens are not generalizable (i.e., can only inform treatment for a specific patient) in isolation due to potentially confounding effects of background gene modifiers. (B) hiPSC cohort clinical trial in a dish (“macromedicine”). Cohorts of bio-banked iPSCs, with various disease associated variants/biomarkers and various genetic backgrounds, are differentiated and cell line to phenotypic recovery outcomes are identified for candidate drugs. Computational analysis is performed to identify externally valid (i.e. results can be used to inform treatment for patients not in cohort) drug specific variant/biomarker to phenotypic recovery correlations. Cohort approaches are ideal for diseases with complex inheritance and access to patient hiPSCs. (C) High-throughput, variant-specific therapeutic screening. Disease associated variants can be introduced into a healthy “parental control line” with scarless genome editing to create an array of isogenic cell lines. Direct assessment of variant/biomarker specific therapeutic response relationships can be observed with high-content screening (or other high-throughput) assays. This approach is ideal for rare monogenic diseases where access to patient-specific hiPSCs is limited.