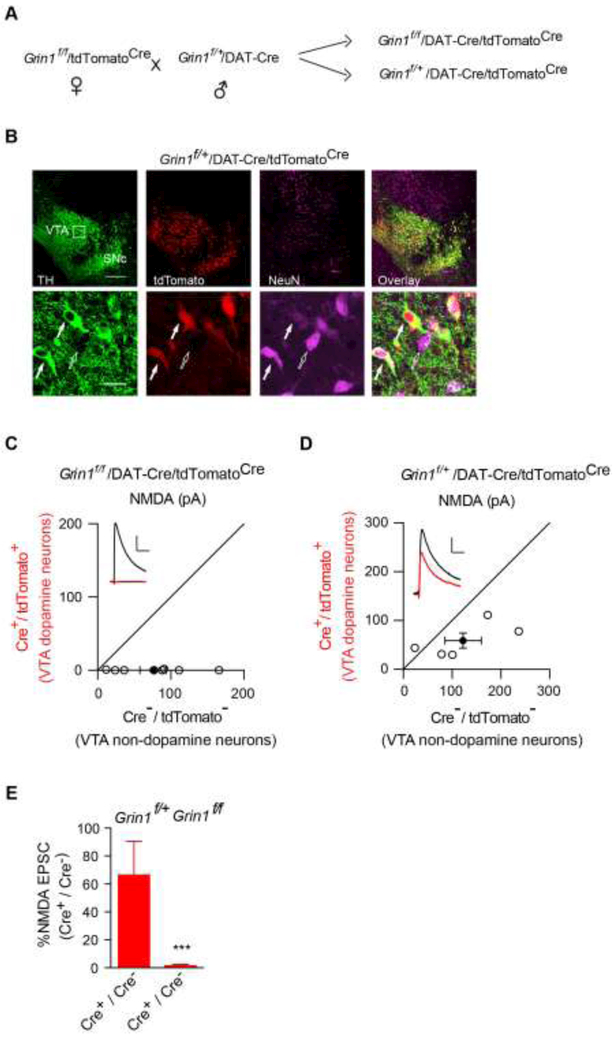
Figure 3. Genetic deletion of NMDARs in VTA dopamine neurons.
(A)Breeding scheme to generate Grin1f/f/DAT-Cre/tdTomatoCre mice.
(B)Immunohistochemical assays in P13 Grin1f/+/DAT-Cre/tdTomatoCre mice confirmed Cre expression selectively in dopaminergic neurons as indicated by complete colocalization of tdTomato, the product of Cre-mediated recombination and tyrosine hydroxylase (TH, green), a cellular marker for dopamine neurons. Note that there were both tdTomato-positive dopamine neurons (solid arrow) and tdTomato-negative nondopamine neurons (open arrow) in the VTA region. NeuN is a neuronal marker (purple). SNc, Substantia nigra pars compacta.
(C-D) Scatter plots and sample traces of NMDA EPSCs in tdTomato-expressing VTA dopamine neurons and nearby tdTomato-negative non-dopamine control neurons from the same slices showed the loss of NMDA EPSCs in dopamine neurons from Grin1f/f/DAT-Cre/tdTomatoCre (C), but not in Grin1f/+/DAT-Cre/tdTomatoCre mice (D).
(E) Bar graph showed normalized NMDA EPSC ratio in dopamine neurons vs nondopamine neurons from Cre+/Cre-in Grin1f/+/DAT-Cre/tdTomatoCremice (66.36 ± 24.02 %, n = 6) and in Grin1f/f/DAT-Cre/tdTomatoCre mice (1.35 ± 1.01 %, n = 9), p < 0.001, t-test. Deletion of the GluN1 subunit abolished NMDA EPSCs in VTA DA neurons from Grin1f/f/DAT-Cre/tdTomatoCre mice. Scale bar: 50 pA and 50 ms.