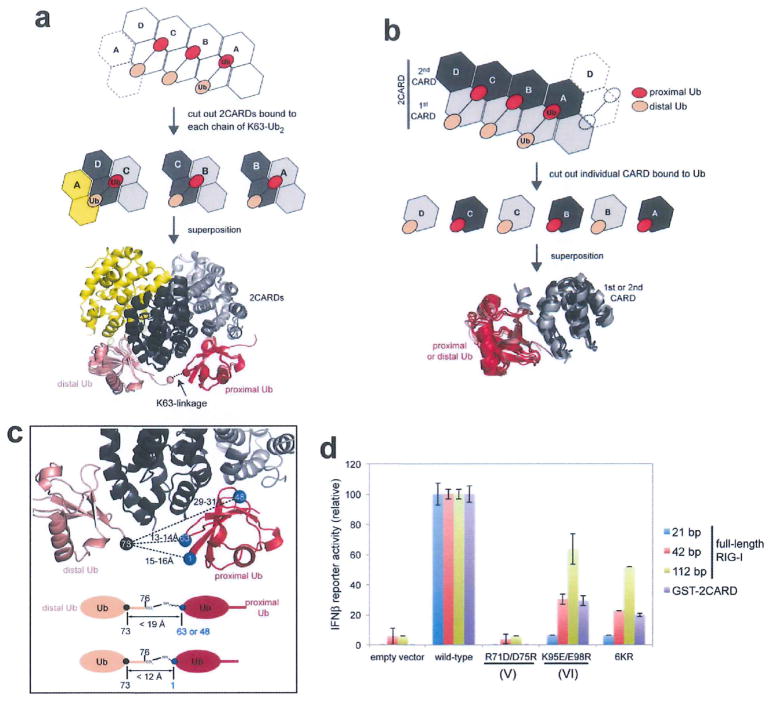
Extended Data Figure 5. Interaction between the 2CARD tetramer and K63-Ub2.
a, Superposition of RIG-I 2CARD in the tetramer (subunits A–D) bound with K63-Ub2. The three Ub2 chains bound to A–B, B–C and C–D–A were superposed by aligning chain A, B and C, respectively, b, Superposition of the first (dark grey) and second (light grey) CARDs from RIG-I 2CARD bound by proximal (red) or distal (salmon) Ubs. The good alignment suggests that the proximal Ub-binding site, IV, in the second CARD is equivalent to the distal Ub-binding site, VI, in the first CARD (see Fig. 3b for the definitions of IV and VI). c, Distance analysis between the C-terminal residue of the distal Ub and K63, K48 and M1 of the proximal Ub. The C-terminal tail (residues 74–76) of the distal Ub is disordered in the structure, and thus the distance was measured between Cα of residue 73 and Cα of the target Lys. Below is the distance requirement for covalent conjugation, assuming 3.5Å spacing per residue in the missing C-terminal tail, 2Å for the C-terminal carboxylate, 6Å for Lys side chains and 1.5Å for N-terminal amine. These distance requirements are satisfied only with K63, thus rationalizing the observed specificity of 2CARD for the K63-linkage. d, IFN-β reporter activity of full-length RIG-I stimulated with 21 bp, 42 bp and 112 bp dsRNAs or isolated 2CARD fused to GST (GST–2CARD) without RNA. Activities were compared between wild type and mutants defective in Ub binding (R71D/D75R and K95E/E98R) or conjugation (6KR), and were normalized against the wild-type values (mean ± s.d., n = 3). 6KR indicates Arg mutation of six Lys (K99, K169, K172, K181, K190 and K193) that are known to be conjugated with K63-Ubn. The signalling activity of full-length RIG-I with 21 bp dsRNA was completely abrogated by R71D/D75R, K95E/E98R or 6KR, suggesting the importance of both Ub-binding and conjugation. The negative impacts of K95E/E98R and 6KR were progressively alleviated by stimulation with increasing length of dsRNA (42 and 112 bp). This restoration of the signalling activity by longer dsRNAs is consistent with our previous report that filament formation of RIG-I on long dsRNA (>60 bp) promotes 2CARD tetramerization by the ‘proximity-induced’ mechanism12. Note that 21 bp, 42 bp and 112 bp dsRNA can accommodate 1–2, 3, 8 RIG-I molecules, respectively. The negative impact of R71D/D75R could not be alleviated by stimulation with longer dsRNAs, which is somewhat at odds with the result with another Ub-binding deficient mutant, K95E/E98R. It is possible that R71D/D75R has more severe defects (in Ub binding or perhaps in 2CARD structure), which could not be overcome by Ub-conjugation or filament formation. For comparison, we also used GST-2CARD, which has been widely used in previous studies1–3. Despite the fact that GST-2CARD cannot form filaments, its sensitivity to the mutations was equivalent to full-length RIG-I with 42 bp, rather than 21 bp dsRNA. This likely reflects the effect of the fusion partner, GST, which forms a constitutive dimer. Isolated 2CARD without GST is a very poor stimulant of IFN-β, thus could not be used in this study.