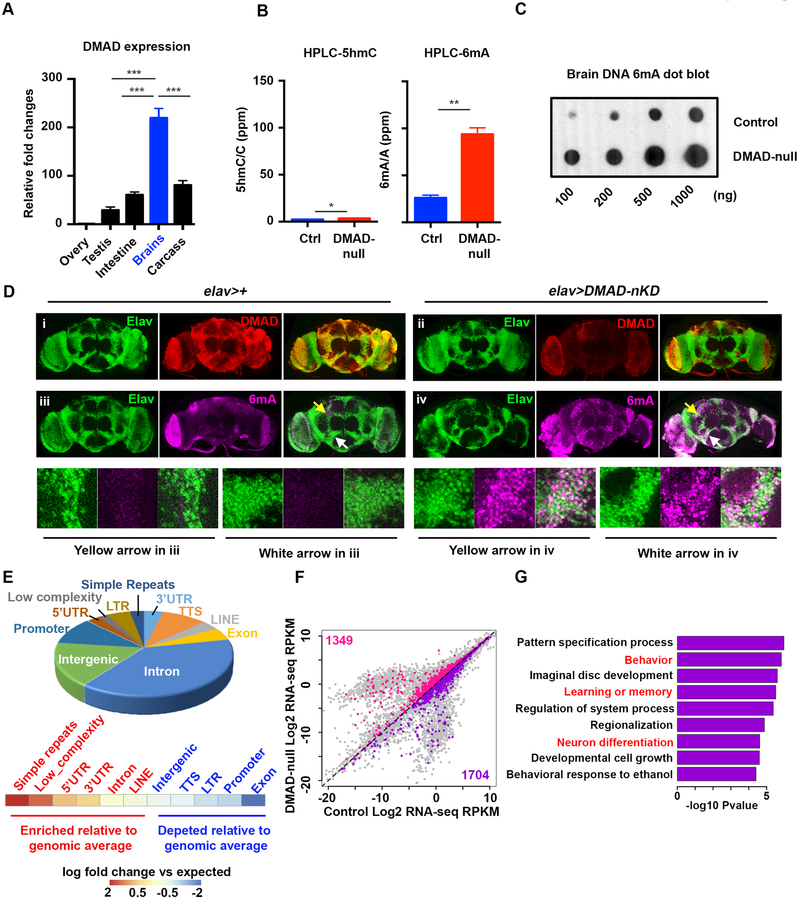
Figure 1. DMAD demethylates intragenic 6mA in Drosophila brains.
(A) qRT-PCR across fly tissues revealed high levels of DMAD expression in heads compared to other somatic tissues (n=3).(B) High resolution HPLC quantification of 5hmC and 6mA in Drosophila brains in the presence (Ctrl) and absence of DMAD (DMAD-null). 5hmC/total C or 6mA/total A are shown as percentage per million nucleotide (ppm). There was a ~3.6-fold increase in 6mA with DMAD depletion while 5hmC was undetectable (n=2).(C) Dot blots using an antibody specific for 6mA confirmed the accumulation of 6mA in DMAD-null fly brains.(D) Confocal images of adult brains stained with anti-Elav (Green), anti-DMAD (Red) and anti-6mA (Purple) in the background of elav-Gal4 alone (i & iii) or in combination with UAS-DMAD miRNA (DMAD-nKD) (ii & iv). Enlarged views of arrowed area in iii and iv are shown below. Compared to control brains (elav-Gal4 alone), DMAD knockdown significantly increased nuclear 6mA levels in elav-expressing cells.(E) Genomic annotation of gain-of-6mA regions in DMAD-null fly brains revealed their intragenic characteristics, A heatmap shows the enrichment of each genomic feature versus expected values.(F) Plot of the global transcriptome in control and DMAD-null fly brains obtained from RNA-seq (n=2). Genes bearing gain-of-6mA regions in their gene bodies were highlighted (up-regulated genes in pink and down-regulated genes in purple).(G) Gene Ontology analysis was performed on a subset of downregulated genes in Figure 1F (purple). Log2 fold change [(KD: WT) < −0.5] was applied as the threshold cut-off. Several biological processes involved in neurodevelopment and neuronal functions were enriched and are highlighted in red. Data are represented as mean ° SEM.