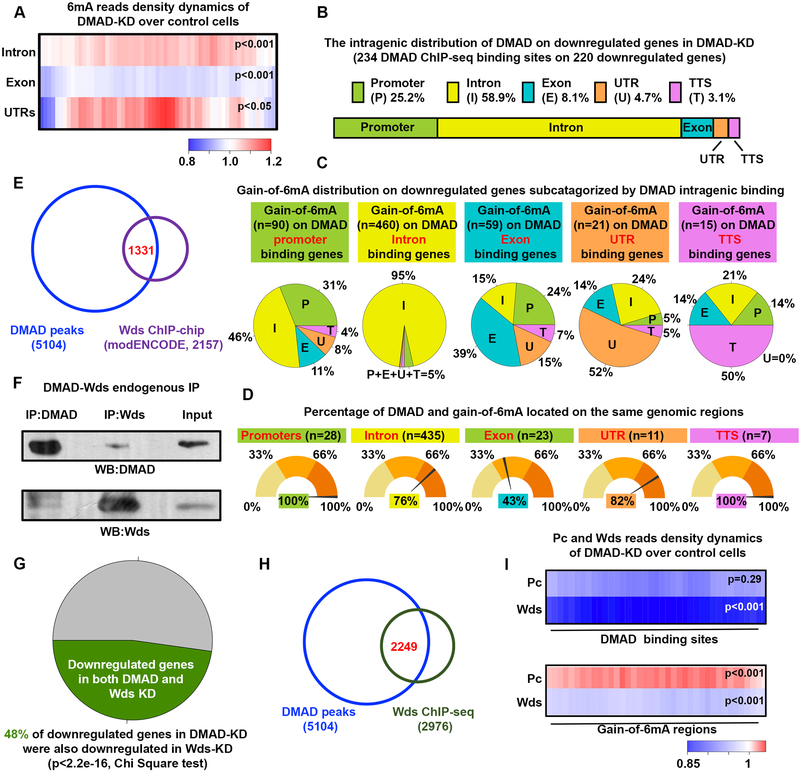
Figure 3. DMAD demethylates intragenic 6mA in cis and coordinates with Trithorax-related protein Wds to regulate gene expression.
(A) Average fold changes in 6mA reads between DMAD-KD and control were calculated for DMAD-occupied gene introns, exons, and untranslated regions. Enrichment or depletion of 6mA on these genomic regions were significant (p<0.001 or p<0.05, Welch Two sample t-tests) (B) Intragenic distributions of DMAD on DMAD-bound downregulated genes bearing accumulation of 6mA upon DMAD knockdown was demonstrated proportionally. DMAD showed strong intronic enrichment on these genes. (C) The genes in (B) were further subcategorized based on the DMAD intragenic association. Gain-of-6mA distributions on each subset of genes were calculated, and the percentages shown in the pie chart. (D) The percentage of genes with both DMAD and gain-of-6mA binding to the same genomic feature were calculated.(E) Venn diagram shows substantial and significant overlap between DMAD and Wds ChIP-chip data (Binomial tests, p<0.001).(F) Co-immunoprecipitation experiments indicated a physical interaction between DMAD and Wds. (G) Downregulated genes in the DMAD-KD cells showed significant overlapping with downregulated genes in the absence of Wds. Chi-square tests were performed. RNA-seq were performed in triplicates. RPKM fold changes <−0.1 were included.(H) Substantial overlap between DMAD and Wds ChIPseq peaks suggested their functional coordination in regulating gene expression.(I) Average fold change in Pc and Wds reads of DMAD-KD over control were calculated for both DMAD-binding sites and gain-of-6mA regions to explore Pc and Wds dynamics in these regions. Heatmap demonstrated a general decrease in both Pc and Wds at DMAD binding sites when DMAD is depleted. A specific and significant increase in Pc binding on gain-of-6mA regions with DMAD depletion was found. Welch Two sample t-tests, p-values were indicated. Data are represented as mean ° SEM.