Fig. 3.
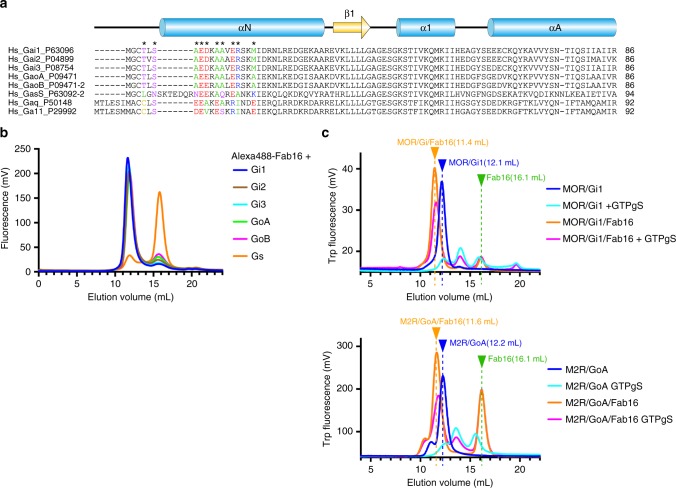
Sequence alignment of G-protein family members and binding profile of Fab16. a Multiple sequence alignment of amino-termini of representative Gα subunits from human. UniProt numbers are provided after each G-protein subtype name. Secondary structures are shown as cylinder (helix) and arrow (strand). The asterisks indicate the residues in contact with scFv16 in Gαi1 and those corresponding residues are coloured according to their property: Positive in blue, negative in red, hydrophobic in green, polar in purple, cysteine in yellow. b Fluorescent SEC analysis of binding of the fluorescently labelled Fab16 with G-protein family members. c Analytical tryptophane fluorescent SEC of μOR/Gi1 and M2R/GoA with GTPγS in the presence or absence of Fab16. Each complex alone runs around 12.2 mL. Upon binding to Fab16, they run at 11.4 mL or 11.6 mL indicating the binding of Fab16 to these GPCR/G-protein complexes. Excess free Fab16 runs at 16.1 mL. Dissociated components upon incubating with GTPγS show smaller peaks at 13.5–16 mL