Figure 6.
FACT Depletion Directly and Indirectly Primes the Transcriptome for Pluripotency
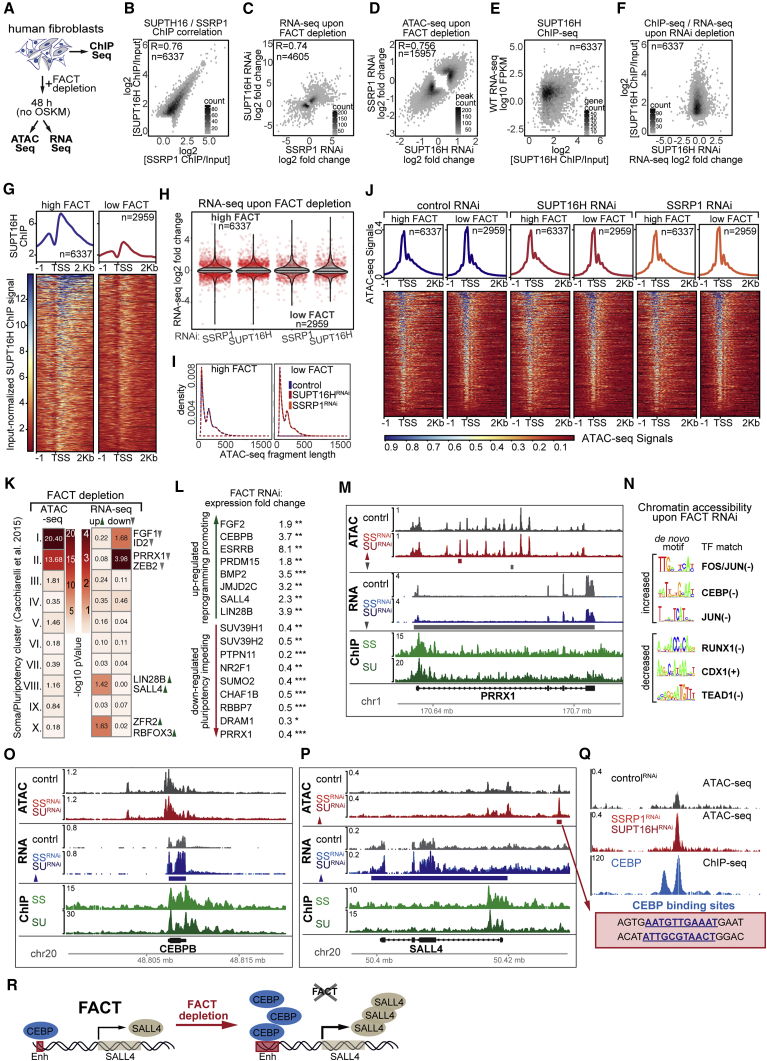
(A) Human fibroblasts were used for ChIP-seq analysis or transfected with siRNAs against human FACT subunits and used for ATAC-seq and RNA-seq analysis 48 hr after knockdown without OSKM induction.
(B) Correlation of SSRP1 and SUPT16H log2-ChIP versus input ratios for TSS windows classified as high for at least one of the two FACT components (see STAR Methods). Density scale reflects number of TSS windows plotted per hexbin. The Spearman correlation Rho value is given.
(C) Correlation of SSRP1 and SUPT16H knockdown RNA-seq log2-fold changes for genes that were significantly changed in at least one of the conditions (see STAR Methods; FDR 0.1). Density scale reflects number of genes plotted per hexbin. The Spearman correlation Rho value is given.
(D) Hexbin density scatterplot of SUPT16H knockdown normalized ATAC-seq log2-fold changes plotted against SSRP1 knockdown normalized ATAC-seq log2-fold changes for ATAC-seq peaks that were significantly differential (FDR 0.01) in at least one of the conditions. Scale shows number of ATAC-seq peaks plotted per hexbin. The Spearman correlation Rho value is given.
(E) Log2 ratio of SUPT16H ChIP versus Input plotted against Fragments Per Kilobase of transcript per Million mapped reads (FPKM) expression values from control RNA-seq for genes classified as FACT high and detected in the RNA-seq. Density scatterplot scale shows number of genes plotted per hexbin.
(F) Log2-fold changes in expression levels from RNA-seq after SUPT16H knockdown plotted against log2 ratio of SUPT16H ChIP versus Input for genes classified as FACT high and detected in the RNA-seq. Density scatterplot scale shows number of genes plotted per hexbin.
(G) Average positional profiles and heatmaps of library- and input-normalized SUPT16H ChIP-seq signal for genes whose TSS windows classified as FACT high or FACT low (see STAR Methods).
(H) Violin plots of RNA-seq log2-fold change distributions following FACT knockdown for genes classified as being FACT high or FACT low. Red jitter dots reflect genes whose expression changed significantly upon FACT knockdown (FDR < 0.1).
(I) Density distributions of control or FACT-knockdown ATAC-seq fragment lengths intersecting TSS windows for genes classified as FACT high or FACT low.
(J) Mean positional profiles and heatmaps of control or FACT-knockdown ATAC-seq signal for genes classified as FACT high or FACT low.
(K) −Log10 enrichment p values of genes assigned ATAC-seq peaks changing, left, or genes showing expression changes upon FACT-knockdown, right, intersecting with clusters of genes previously reported to display specific expression profile changes during OSKM-induced reprogramming (Cacchiarelli et al., 2015). I, early somatic; II, late somatic; III, metabolic processes; IV, late embryogenesis; V, early embryogenesis; VI, pre-implantation; VII, shared soma versus pluripotency; VIII, early pluripotency; IX, late pluripotency; X, neuro-ectoderm.
(L) RNA-seq fold changes upon FACT-knockdown for key genes previously implicated in reprogramming or pluripotency. Stars reflect adjusted p values.
(M) Browser shot of ATAC-seq signal and library-normalized RNA-seq signal upon SSRP1 (SSRNAi) or SUPT16H RNAi (SURNAi), and FACT ChIP-seq signals for the PRRX1 gene. ATAC-seq peaks and RNA-seq genes called as significantly changed are noted below the respective signal tracks.
(N) De novo-generated motifs from ATAC-seq peaks differentially changing upon FACT-knockdown and TF family-representatives matching the given motif.
(O and P) Browser shots as described in (M) for the (O) CEBPB and (P) SALL4 genes.
(Q) Zoomed-in browser shot of upstream enhancer-candidate for SALL4. Shown are control and FACT-knockdown signals for ATAC-seq and publicly available CEBPB ChIP-seq in human embryonic stem cells (ESCs). Two CEBPB biding sites were found within the 387-bp accessible region by matching the consensus RTTKCAYMAY and allowing one mismatch.
(R) Model for how FACT depletion primes for reprogramming: direct changes in gene expression for TFs such as CEBPB lead to promoter distal, indirect chromatin accessibility changes at genomic loci of key regulators such as SALL4.