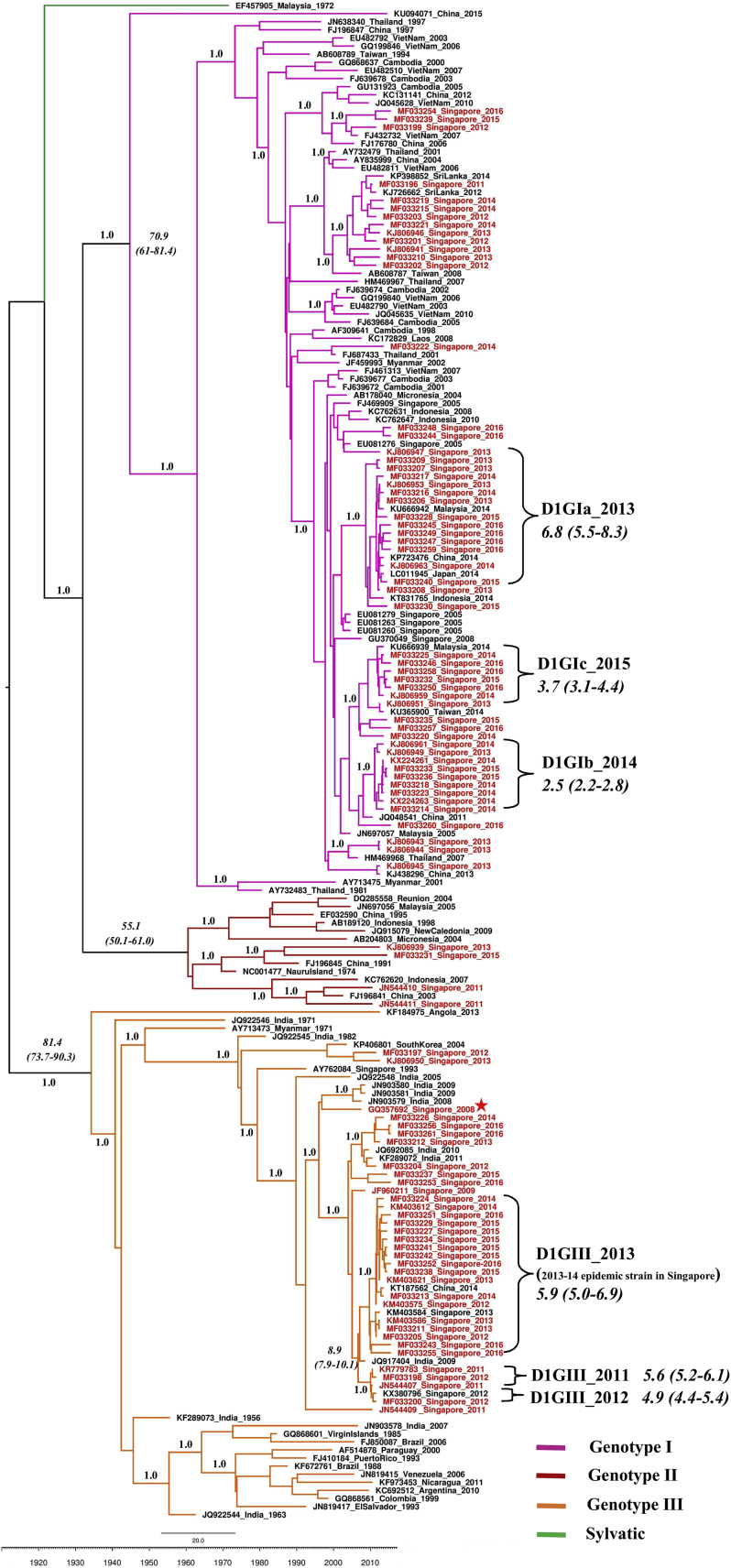
Figure 1.
Phylogenetic and tMRCA Analysis of DENV-1
The time-scaled maximum clade credibility tree was constructed using the Bayesian Markov Chain Monte Carlo (MCMC) method implemented in the BEAST package v1.7.4. The analysis included 93 complete polyprotein sequences selected to cover all possible viral genetic diversity observed during this study (highlighted in red) and 94 sequences retrieved from GenBank database. Each genotype is shown in different colors as per the tree legend. The local strain (imported case from India) that clustered in the outgroup of genotype III lineages is shown with an asterisk. Numbers on branches represent the posterior probability values. The tMRCA values in years are shown in italics, with 95% highest posterior density (HPD) values in brackets. The scale bar shown above the time scale is substitutions/site/year.