Figure 1.
IFN-Stimulated ISGF3 Recruitment and Transcriptional Activity
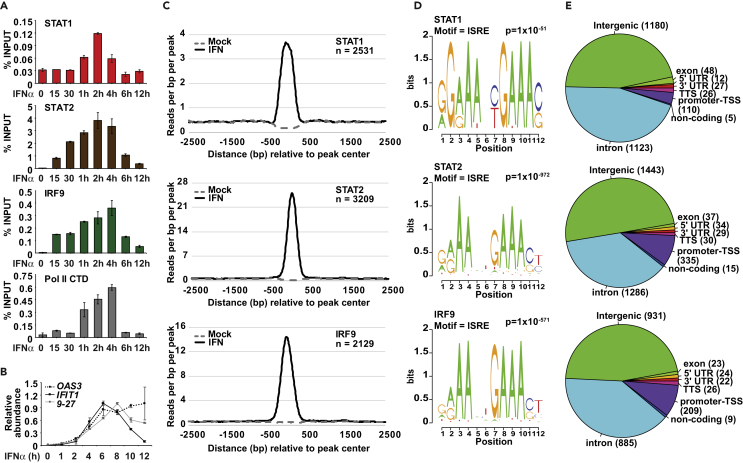
(A) ChIP analysis of IFNα-induced STAT1, STAT2, IRF9, and Pol II C-terminal domain (CTD) recruitment at the OAS3 promoter locus in HeLa cells after mock treatment (0 min) or IFNα stimulation for 15 min, 30 min, 1 hr, 2 hr, 4 hr, 6 hr, and 12 hr. Error bars denote mean ± SD of three technical replicates.
(B) Gene expression analysis of OAS3, IFIT1/ISG56, and IFITM1/9–27 after mock treatment (0 hr) or 1-, 2-, 4-, 6-, 8-, 10-, or 12-hr IFNα treatment. Relative abundance is normalized to GAPDH. Error bars denote mean ± SD of three technical replicates.
(C) Normalized sequencing tag density of mock-treated (dashed) and IFNα-stimulated (solid) STAT1 (top), STAT2 (middle), and IRF9 (bottom) binding at 2,531, 3,209 and 2,129 genomic loci representing sites with a ≥ 2-fold increase in occupancy after IFNα treatment. Tag density is computed 2,500 bp upstream and downstream of the peak center and is grouped into 10 bp bins.
(D) DNA sequence logo of the most frequent de novo motif identified from 2,531 STAT1 peaks (top), 3,209 STAT2 peaks (middle), and 2,129 IRF9 peaks (bottom) as described in Table S1. For each position, the sequence logo bit height corresponds to its relative frequency within the sequence. The associated motif name and p value are identified above the logo.
(E) Distribution of specific annotated DNA (intergenic, intron, promoter-TSS, exon, 5′ UTR, 3′ UTR, non-coding) and the corresponding number of peaks from 2,531 STAT1 peaks (top), 3,209 STAT2 peaks (middle), and 2,129 IRF9 peaks (bottom).
See also Table S1.