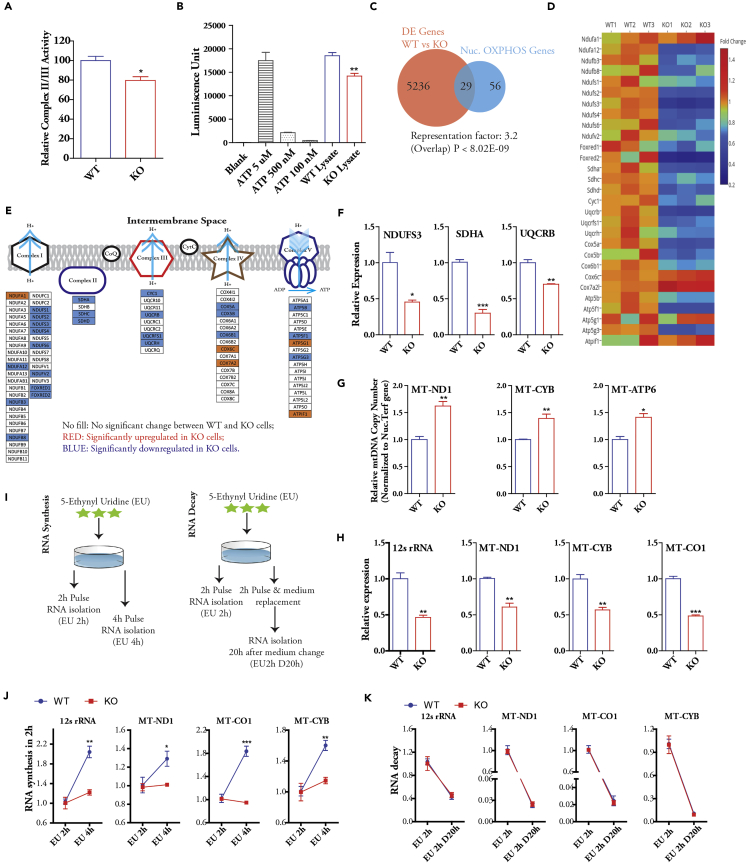
Figure 2.
OXPHOS Defects Are Linked to Impaired mtDNA and Nuclear OXPHOS Gene Expression in β-Actin Knockouts
(A) Mito complex II/III activity assay of isolated mitochondria; n = 3 independent experiments.
(B) Cellular ATP level determination; n = 3 independent experiments.
(C) Venn diagram showing significant overlap of differentially expressed genes in WT versus KO MEFs and OXPHOS genes encoded by nucleus: Fisher's exact test.
(D) Relative expression levels of nuclear OXPHOS genes differentially expressed between WT and KO cells by RNA-seq. The mean value of WT samples was set as 1.
(E) Distribution of differentially expressed nuclear OXPHOS genes.
(F) Quantification of mRNA level of Ndufs3, Sdha, and Uqcrb genes by qPCR; n = 3 biological replicates.
(G) mtDNA level determination using MT-ND1, MT-CYB, and MT-ATP6 genes by qPCR; n = 3 independent experiments.
(H) Transcript level of 12s rRNA, MT-ND1, MT-CYB, and MT-CO1 genes by qPCR; n = 3 independent experiments.
(I–K) Schematics of RNA synthesis and RNA decay experiments are shown in (I). qPCR analysis of mt-RNA synthesis (J) and decay (K); n = 3 biological replicates.
Data are presented as mean ± SEM *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. Student's t-test. SEM, standard error of the mean.
See also Figure S2.