Figure 6.
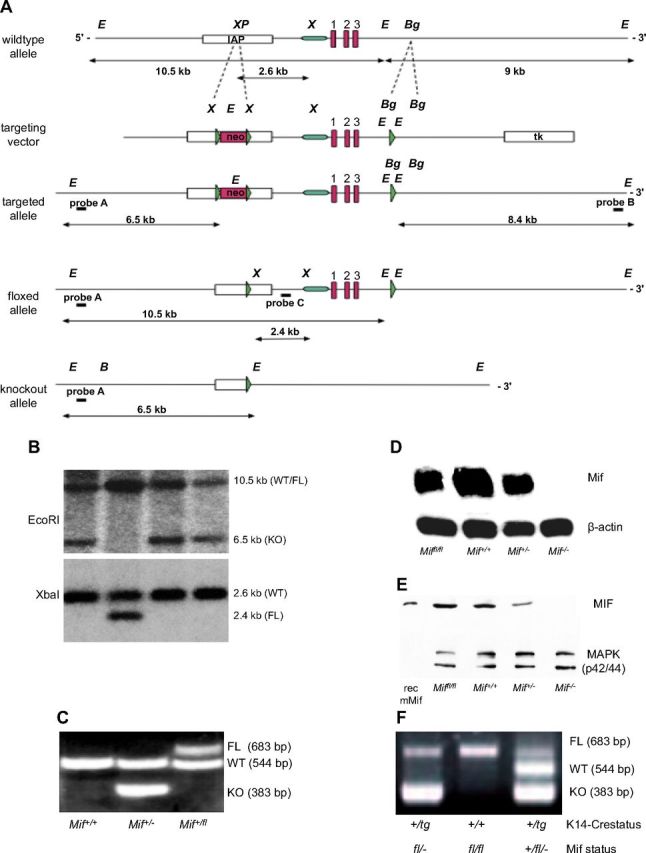
Absence of epidermal MIF, but not of MIF receptors CD74 and CD44, reproduces the phenotype of the complete Mif knockout. A) Design and initial targeting of C57Bl/6 ES cells is reported in the previous work (14). After transient Cre-expression in targeted ES cells, in vitro clones, ES cell clones with floxed Mif (FL), and deletion of the neo selection cassette were identified by sensitivity to GA101. Neo-deleted, floxed MIF ES cells were injected into BALB/c blastocysts and resulting chimeras were bred to C57Bl/6 mice to obtain pure C57Bl/6 Miffl/+ mice. Homozygous Miffl/fl females were crossed with K14-Cre transgenic males to achieve epidermis-specific K14-Cre+/tg; Mif−/− mice. B) Selection of Miffl/fl ES cells after transient transfection with a Cre-expressing plasmid by restriction fragment length polymorphism of EcoRI (top) and XbaI (bottom) digested genomic DNA and southern blotting with an external/internal probe. EcoRI-digest and external probe A in G418-sensitive clones: 10.5-kb band indicates the WT or floxed allele, 6.5-kb indicates deletion of both Mif and Neo. XbaI-digest and internal probe C: 2.6-kb band indicates WT allele, 2.4-kb band corresponds to floxed allele. C) Representative example of the genotyping PCR differentiating Mif+/+, Mif+/−, and Mif+/fl mice. D) Northern blotting for MIF mRNA from skin of Miffl/fl, Mif+/+, Mif+/−, and Mif−/− mice. β-Actin mRNA used as loading control. E) Western blotting for MIF protein from skin of Miffl/fl, Mif+/+, Mif+/−, and Mif−/− mice. MAPK used as loading control. F) Representative example of the genotyping PCR from tail DNA in K14-Cre transgenic and K14-Cre-negative Miffl/fl mice. Presence of the K14-driven Cre recombinase leads to deletion of the mif gene in keratinocytes visualized by the presence of the 383-bp band.
