Summary
A clustered regularly interspaced short palindromic repeats (CRISPR)-like “mimivirus virophage resistance element” (MIMIVIRE) system, which contains specific cascade genes and a CRISPR array against virophages, was reported in mimiviruses. An essential component of the MIMIVIRE system is R354, encoding a nuclease and a likely functional homolog of Cas4. Here we show that R354 is a dual nuclease with both exonuclease and endonuclease activities. Structural analysis revealed that the catalytic core domain of R354 is similar to those of Cas4 and λ exonuclease despite their low sequence identity. R354 forms a homodimer that is important for its exonuclease but not endonuclease activity. Structural comparisons between the active and semi-active states of R354 demonstrated that an activation loop adjacent to the catalytic site is critical for enzymatic activity. Overall, the results suggest that R354 belongs to a novel MIMIVIRE system involved in innate virus immunity and provides a template for the identification of new CRISPR systems in other species.
Subject Areas: Virology, Structural Biology, Protein Structure Aspects
Graphical Abstract
Highlights
-
•
The key component of Cas4-like R354 in MIMIVIRE is a dual nuclease
-
•
Structural insights into R354 indicate that it is fully active as a homodimer
-
•
The semi-active state of R354 is a monomer
Virology; Structural Biology; Protein Structure Aspects
Introduction
The clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) system is a unique adaptive immunity mechanism of bacteria and archaea that mediates defense against invasion by foreign microbes (Wiedenheft et al., 2012). Currently, six types of CRISPR-Cas systems (I–VI) have been identified (Hudaiberdiev et al., 2017, Koonin et al., 2017, Makarova et al., 2015). Among them, the type I, II, and V systems consist of several subtype-specific genes (A–D and U of type I; B and C variants of type II, and A, B, and E of type V) (Koonin et al., 2017). These systems also include a variety of Cas proteins, including Cas4, which form complexes that mediate the recognition of foreign DNA sequences (Brouns et al., 2008, Sorek et al., 2013).
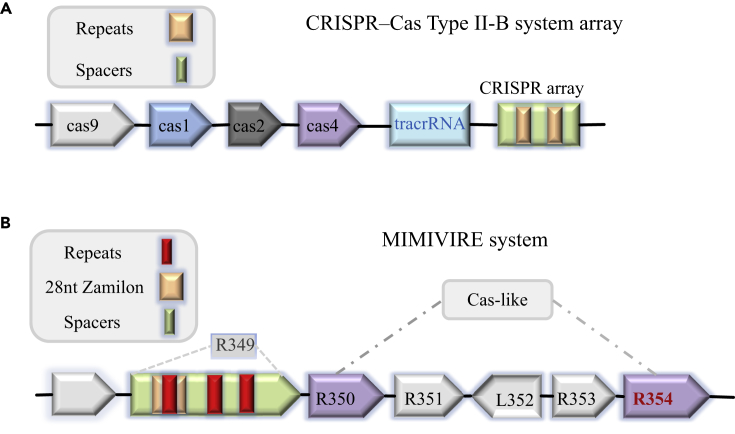
Notably, in a recent study, a CRISPR-like mimivirus virophage resistance element (MIMIVIRE) system, which consists of a cascade of genes and a CRISPR array, was reported to protect mimiviruses against virophages (Levasseur et al., 2016) (Figure 1). Specifically, this system functions similarly to the Cas proteins and interferes with virophage invasion (Levasseur et al., 2016) (Figure 1). The nuclease R354, a putatively functional homolog of Cas4 involved in spacer processing for adaption to foreign DNA (Koonin et al., 2017, Plagens et al., 2012, Seed et al., 2013), was found to be essential for MIMIVIRE function.
Figure 1.
The MIMIVIRE Defense System
(A) Schematic representation showing the prokaryotic CRISPR-Cas system, which is representative of the type II-B system; all components are highlighted in different colors (Hudaiberdiev et al., 2017, Koonin et al., 2017).
(B) Schematic representation showing the viral MIMIVIRE system. Zamilon, a novel virophage, was reported to infect giant viruses from the Mimiviridae family. A specific 28-nucleotide-long sequence in some mimivirus genomes was defined to be identical to the fragment DNA sequence of Zamilon; this pattern is similar to the CRISPR-Cas system, and the unique sequence was named Zamilon. The inserted 28-nucleotide-long sequence is AATCTGATAATGAATCTGATAATGAATC, and the derived 15-nucleotide repeated unit is TGATAATGAATCTGA (Brouns et al., 2008, Gaia et al., 2014).
Although structural information on Cas4 proteins is available, the mechanisms underlying R354-mediated function remain elusive. Here we present the crystal structures of R354 in the active and semi-active states at a resolution of 2.8 Å and 3.0 Å, respectively. The protein is composed of a central channel and a lateral groove, suggesting the entry and exit of the substrate in the cleavage process. Importantly, the crystal structure in the dimerization state, together with biochemical analyses, demonstrates that R354 homodimerization is an exclusive requirement for the exonuclease in double-stranded DNA (dsDNA) but not for the endonuclease activity, revealing a unique regulatory mechanism for R354 function. Altogether, functional and structural analyses suggest that R354 is a functional Cas4-like protein of the viral MIMIVIRE system with a unique regulatory mechanism. Our findings also provide insights into the evolution of nucleases, which might enable the identification of new CRISPR systems in other species.
Results
R354 Is a Dual Nuclease
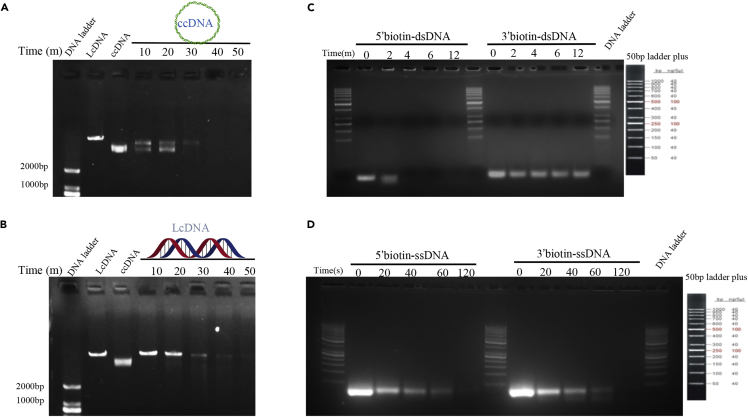
We investigated the enzymatic activity of R354 using purified protein, as a first step toward the characterization of this MIMIVIRE system protein (Figure S1). We selected plasmid pET21b-LipL as the general substrate for determining R354 endonuclease activity after a digestion-based endonuclease activity assay (Figure 2A). Furthermore, digestion of linear and circular double-stranded plasmid DNA was performed for examining R354 exonuclease activity; the substrate disappeared with time (Figures 2A and 2B). Because most nucleases are divalent metal-dependent enzymes, we performed nuclease assays with different metals. As expected, the nuclease activity of R354 was maximal in the presence of Mg2+, Mn2+, Fe2+, or Zn2+, but not Ca2+ (Figure S2), which is consistent with the results of previous reports (Lemak et al., 2013, Lemak et al., 2014).
Figure 2.
R354 Is a Dual Nuclease
(A) Characterization of the endonuclease activity of R354 by an endonuclease activity assay in vitro. ccDNA refers to circular coiled-coil DNA, LcDNA refers to linear coiled-coil DNA.
(B) Characterization of the exonuclease activity of R354 by an exonuclease assay in vitro.
(C) Exonuclease activity assay using biotin-labeled dsDNA. R354 was incubated with the labeled dsDNA with blunt ends or 5′ or 3′ overhangs, and the reactions were analyzed by agarose gel electrophoresis (AGE). Results indicate cleavage of the blunt end and the 3′ overhang of the dsDNA.
(D) Exonuclease activity assay using biotin-labeled ssDNA. R354 was incubated with the labeled ssDNA with blunt ends or 5′ or 3′ overhangs, and the reactions were analyzed by AGE. The results indicated cleavage of the blunt ends and the 5′ and 3′ overhangs of the ssDNA. The experiments were repeated three times.
In addition, R354 cleaved a 5′ or 3′ labeled biotinylated dsDNA substrate in the 3'→5′ direction in the exonuclease activity assay (Figure 2C). Conversely, R354 cleaved single-stranded DNA (ssDNA) in both 3'→5′ and 5'→3′ directions (Figure 2D), which is consistent with the function of the Cas4 protein SSO1391 (Lemak et al., 2013). We thus consider the dual nuclease R354 to likely be a functional Cas4 protein in the MIMIVIRE system. Although our data suggest that R354 functions as a dual nuclease, these results are preliminary and further characterization is required to identify the exact mechanism of action.
General Structure of R354
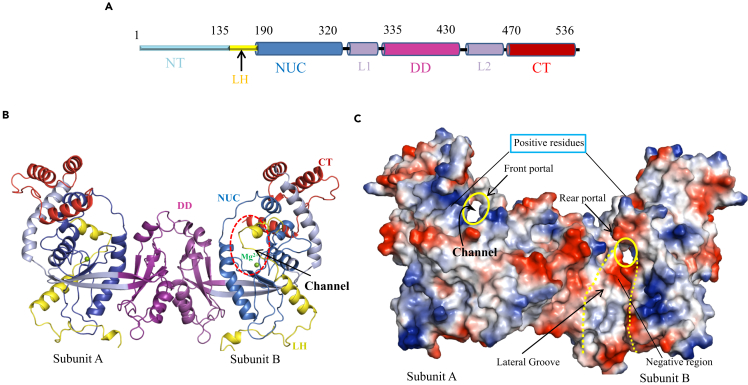
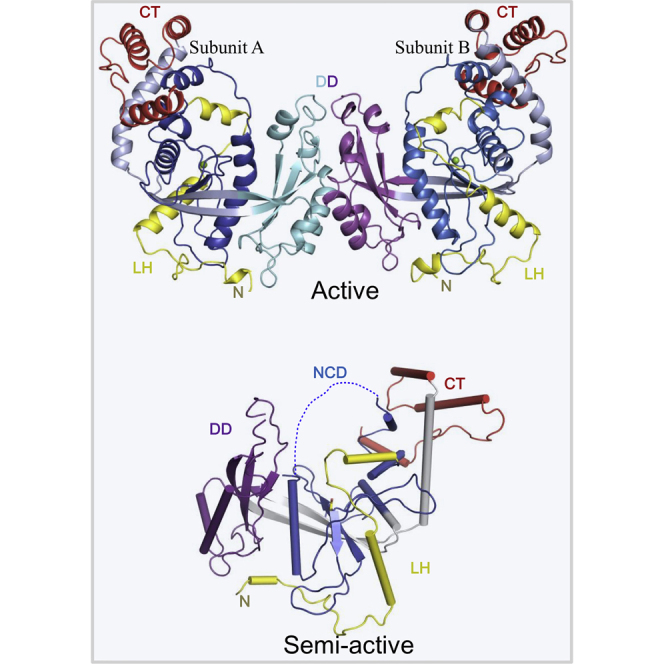
We determined the crystal structures of the wild-type and N-terminal deletion (NTD) mutant of R354 (deletion of amino acids 1–128) at 3.0 Å and 2.8 Å, respectively. Nuclease activity assays showed that the NTD deletion mutant does not affect the activity (Figures 2A and S3). Notably, the structures are also identical; the N-terminal fragment (residues1-137) was not observed in the density maps of the wild-type structure. However, because the structure of Del1-128 was determined at higher resolution than that of the wild-type protein, we only discuss the NTD deletion structure for simplicity (Table S1). The protein consists of two subunits (A and B) that form an asymmetric unit (Figure 3B and Table S1). Each subunit is composed of an N-terminus (NT129-194), a nuclease region (NUC), a dimerization domain (DD), and a C-terminus (CT; Figure 3A). The NT is connected to the NUC through a loop-linked helix (LH), and two unique linkers, L1 and L2, bridge the NUC and DD and the DD and CT, respectively. The LH-spanning residues 135–190 (LH135–190), which are located at the NT, contain a helix located in the middle of a large loop. DD335–430 and CT470–536 surround the core structure of NUC192–320, which presumably maintains R354 in its functional fold. Overall, the structure of R354 includes a five-stranded β sheet and 16 α helices, which form a central channel and a lateral groove (Figure 3B). The central channel contains a front portal, tunnel, and rear portal, which are possibly used for DNA entry, passage, and exit, respectively (Figure 3C). Several positively charged residues were observed at the front and rear portals of the channel. In contrast, the tunnel harbored several negatively charged residues, suggesting that the protein interacts with polar substrates.
Figure 3.
Overall Structure of R354
(A) Graphic depictions of R354 domain organization.
(B) The overall structure of R354; Mg2+ is highlighted in green sphere.
(C) Surface representation of R354. The front and rear portals are both highlighted with yellow circles; the lateral groove is indicated by dashed lines.
R354 in Active and Semi-active States
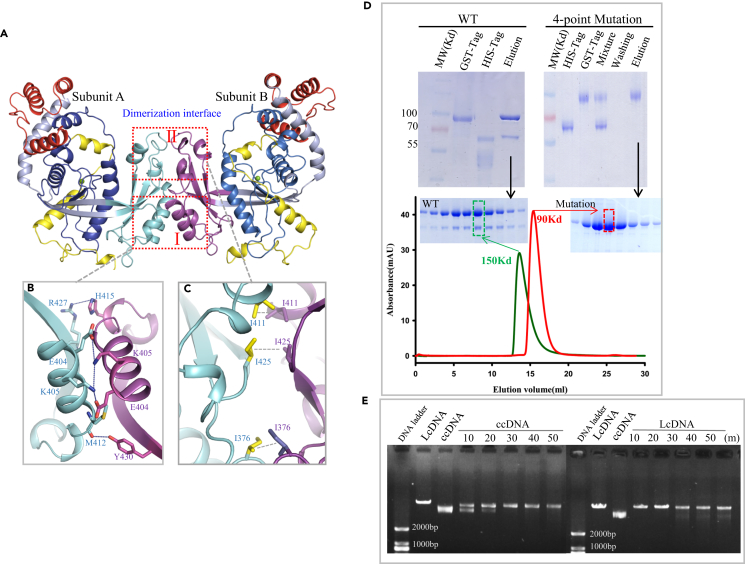
Crystallographic analysis of R354 packing revealed that it forms a very tight protein dimer (Figure 4A). Analysis of the dimer interface indicated that protein-protein interactions are mediated by the Lys405 and Glu404 residues of subunits A and B, respectively, which form a strong interaction at the interface (Figure 4B). A clear interaction was also observed between the backbone carbonyl of Met412 of subunit A and the side chain of Tyr430 of subunit B (Figure 4B). In addition, an interaction was formed by Arg427 of subunit A and His415 of subunit B. Notably, other prominent hydrophobic contacts existed among the Leu376, Ile411, and Ile425 residues of each subunit (Figure 4C). These interactions were observed in the structures of both the wild-type protein and the NTD mutant, which demonstrated that the N-terminal 128 residues are not required for dimerization.
Figure 4.
Dimerization of R354
(A) Dimerization surface I and surface II formed by the interaction of subunit A and subunit B of R354.
(B) Close up of the interactions of interface I. Hydrogen bonds and ionic bonds are indicated by blue dashed lines.
(C) Close up of the interactions of interface II. The hydrophobic interactions are indicated by gray dashed lines.
(D) Verification of dimerization of R354 in solution by pull-down assay and gel filtration analyses; details described in Transparent Methods.
(E) Verification of the dimer-dependent exonuclease activity of R354 using a variant harboring four point mutations (E404A, K405A, M412A, and R427A). LcDNA refers to linear coiled-coil DNA; ccDNA refers to circular coiled-coil DNA.
Next, we performed biochemical assays to determine whether R354 exists as a dimer in solution. Pull-down and gel filtration assays showed that R354 exists as a dimer (Figures 4D–4F), whereas binding and nuclease activity assays using an R354 variant harboring four amino acid substitutions, namely, Glu404Ala, Lys405Ala, Met412Ala, and Arg427Ala, showed that dimerization was critical for R354 exonuclease activity in both dsDNA and ssDNA (Figures 4E and S4).
Our results show that R354 is a functional homodimer. Furthermore, R354 dimerization provides physical interactions for enhancing the activation of exonuclease function. However, further studies are required to elucidate the mechanism of R354 activation.
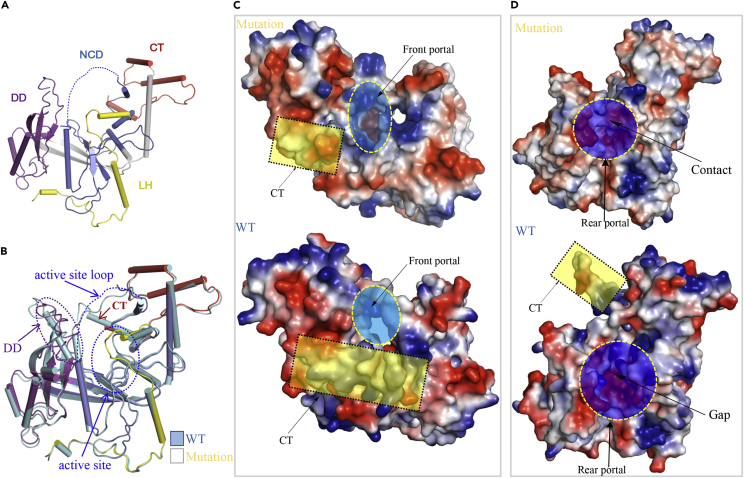
We therefore successfully determined the crystal structure of an R354 variant harboring four amino acid substitutions (Glu404Ala, Lys405Ala, Met412Ala, and Arg427Ala) that is in a monomeric state, as supported by pull-down assays (Figure 4D). The monomeric structure is useful for investigating the mechanisms underlying R354 activation (Figure 5A and Table S1). Structural differences between the dimer and monomer were observed (Figure 5B). Specifically, the active site loop (Cys215–His229) was well ordered in the dimer but not observed in the density maps of the monomeric structure. Notably, the tunnel at the active site could not be formed well in the monomer, as shown in Figure 5C. In addition to the conformational changes at the active site, prominent changes were also observed at the dimer interaction interface. Importantly, the large loop (Ser371–Lys389) was flexible in the mutant, and the lengths of certain β sheets and α helices were partially altered (Figure 5B). Moreover, the C-terminal portion of the CT (residues520–538) was partially disordered and it did not form the helix observed in the monomeric state (Figures 5B–5D). The importance of this region was verified by enzymatic activity assays (Figure S5). Overall, these observations prompted us to hypothesize that the dimerization state is important for enhancing the activation of R354 exonuclease function.
Figure 5.
The Semi-active State of R354
(A) Overall structure of R354 harboring the four point mutations (E404A, K405A, M412A, and R427A). The active site loop region indicated by the dotted line, which is not observed in the electron density map and the electron density map for the C terminus, is also not observed.
(B) Structural comparisons of the active and semi-active states of R354. The active state is more rigid than the semi-active state, particularly in the portions DD, NCD, and CT.
(C) The electrostatic representations viewed from the front portal for both semi-active and active states, the different front portals, and the CT portions are highlighted in blue and yellow colors, respectively.
(D) The electrostatic representations viewed from the rear portal for both semi-active and active states. The different rear portals and the CT portions are highlighted in green and yellow colors, respectively.
Based on these conformational observations, we posit that dimerization is required for complete activation of R354. The R354 monomer was loosely folded, highlighting that the rigid fold is required for the full exonuclease activity of R354.
Structural Analysis of the Active Site
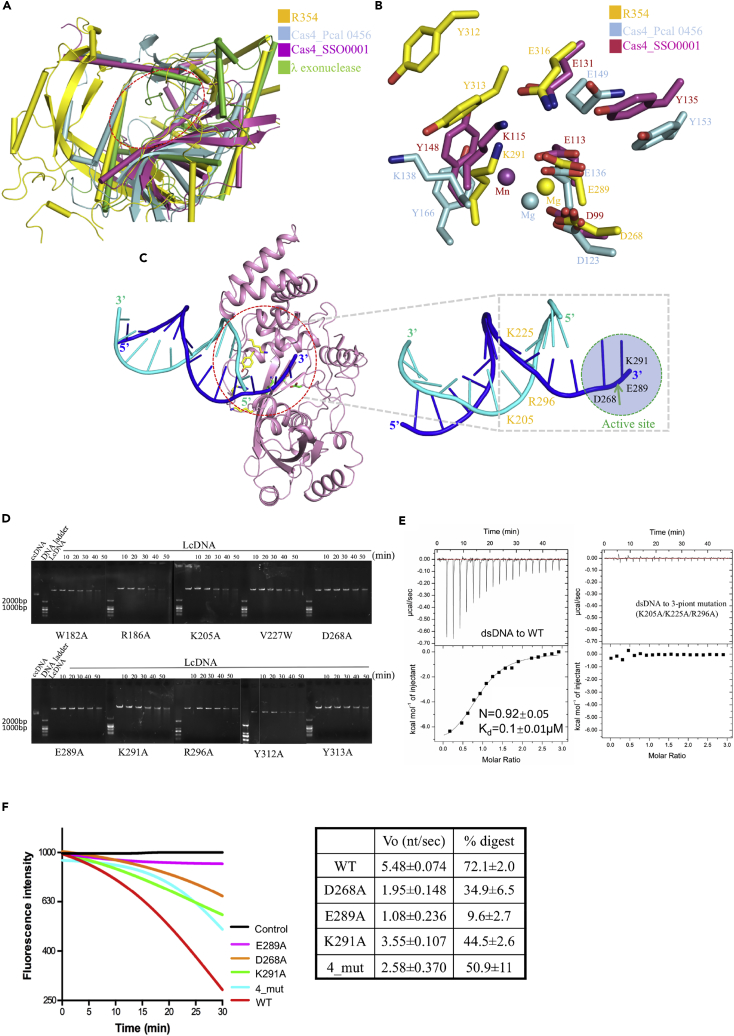
A Dali search identified Cas4 protein SSO0001 (Lemak et al., 2013) (PDB ID is 4ic1 with a root-mean-square deviation [RMSD] of 3.4 Å for 162 equivalent Cα atoms), Pcal 0546 (Lemak et al., 2014) (PDB ID is 4r5q with an RMSD of 4.2 Å for 143 equivalent Cα atoms), and λ exonuclease (Kovall and Matthews, 1997, Zhang et al., 2011) (PDB ID is 3sm4 with an RMSD of 2.8 Å for 186 equivalent Cα atoms) as being structurally similar to R354. This structural similarity was largely attributed to the nuclease domain (NC186_346) despite low levels of sequence identity (Figures 6A and S6). Together, these findings indicate that the CRISPR system could have evolved in species other than bacteria and archaea, including viruses. Moreover, the findings suggest that the nuclease and CRISPR systems of bacteria and mimiviruses evolved from a cenancestor and that the R354 coding sequence was acquired via vertical inheritance (Figure S7). Therefore we can interpret the catalytic mechanism of R354 by structural comparison.
Figure 6.
Structural Analysis
(A) Structural comparisons of R354, Cas4_Pcal 0456, Cas4_SSO0001, and λ exonuclease. The similarities between the core catalytic domains of the four proteins are highlighted by red dashed lines.
(B) Comparison of active sites of R354 and Cas4; some residues are conservative in both active sites, which suggests that R354 has similar function to Cas4.
(C) A cartoon presentation of the dsDNA modeled into the structure of R354 (blue for the cleaved strand with the 3′-OH and cyan for the other strand), which shows interactions between DNA and R354. DNA was modeled by superimposing the catalytic core domain of R354 and λ exonuclease (PDB ID 3SM4Zhang et al., 2011). The red dotted ring is interpreted in the right panel. Residues putatively interacting with the DNA substrate and involved in catalytic reaction are shown as sticks in the left panel. The cut site of DNA strand and the active site of R354 are indicated with highlighted label; the key residues are labeled in the right panel.
(D) Verification of key residues involved in R354 enzymatic activity using activity assays. LcDNA refers to linear coiled-coil DNA; ccDNA refers to circular coiled-coil DNA.
(E) Isothermal titration calorimetry (ITC) measurement of dsDNA binding. Representative ITC raw data and binding trace for dsDNA titrated into the solution containing wild-type (WT) R354 in the left panel: Kd = 0.1 ± 0.01 μM; N = 0.92 ± 0.05. The variant harboring three point mutations (K205A/K22A/R296A) can disrupt the dsDNA binding in the right panel.
(F) Quantitative exonuclease assays. (Left) Comparison of exonuclease activities of wild-type and representative variants of R354. The reactions began to record (m = 0) at the signal stabilized by the addition of enzyme to reaction buffer. (Right) The reaction parameters are shown in the table. The first column indicates the rate determined by the initial slope of the reaction. The percent of DNA substrate digested at 30 min is shown in the second column by comparison of detected fluorescence relative to reactions between positive (WT) and negative controls. Data are measured with three independent replicates for standard errors.
In contrast to Cas4 and λ exonucleases, which require a toroidal state for their exonuclease activities, dimerization was essential for the exonuclease but not the endonuclease activity of R354. These findings suggest that R354 may employ a unique regulatory mechanism. However, the interactions between Mg2+ and residues Asp268, Glu289, and Lys291 within the active site of R354 were similar to those of Mn2+, Asp99, Glu113, and Lys115 in Cas4 SSO0001 and Mg2+, Asp123, Glu136, and Lys138 in Cas4 Pcal 0546 (Figure 6B). These three active site residues are critical for protein function, and a single mutation can almost abolish exonuclease activity. In addition, other critical residues, including Gln316, Tyr313, and Tyr312, were also similar to Glu131, Tyr135, and Tyr148 in Cas4 SSO0001 and Glu149, Tyr153, and Tyr166 in Cas4 Pcal 0546 (Figure 6B); these three residues are likely associated with substrate passage through the central channel. The Mg2+ in the catalytic site possibly enhances the catalytic rate, although the detailed mechanism is not fully understood. Possibly, metal ions coordinate the oxygen atom to trigger a nucleophilic attack and form the leaving group during cleavage of the scissile phosphodiester bond (Horton and Perona, 2001, Steitz and Steitz, 1993). The structural and functional similarities and the presence of conserved residues thus suggest that R354 is a Cas4-like protein.
To reveal the interaction of DNA with R354, we superimposed the structures of λ exonuclease in complex with dsDNA (Zhang et al., 2011) (PDB ID: 3SM4) on our R354 structure, resulting in a good fit between DNA from the λ exonuclease structure and the surface of R354 (Figure 6C). The modeling indicated obviously that R354 has two groups of residues for DNA binding and digesting, which are located at the front portal and inner tunnel (Figures 3C and 6C), respectively; some other residues adjacent to the active site are possibly involved in DNA moving. To further verify the importance of these residues, we mutated the residues within the central channel, including those at the entry and exit sites, to alanine and performed enzymatic activity assays with the mutants. The residues at the front portal site are probably involved in contacting the downstream portion of the dsDNA for entering the tunnel (Arg205, Arg296, and Tyr313), residues at the hydrophobic wedge may be required for DNA movement (Val227), the active site in the central channel is required for catalysis (Asp268, Glu289, and Lys291), and the residues at the rear portal site and slide groove are possibly used for phosphate exit and sliding out of the processed ssDNA (His145, Arg171, Gln177, Trp182, and Arg186). Substitution of each of the above residues adjacent to the active site with alanines (Trp182Ala, Arg186Ala, Lys205Ala, Val227Ala, Asp268Ala, Glu289Ala, Lys291Ala, Arg296Ala, Tyr312Ala, and Tyr313Ala) nearly abolished the exonuclease activity of R354 (Figure 6D), highlighting their functional significance. In addition, isothermal titration calorimetry (ITC) measurements of some variants (Arg205Ala, Arg296Ala, and Tyr313Ala) supported the proposed binding sites for dsDNA (Figure 6E). Moreover, a quantitative exonuclease assay was performed for the representative residues with the corresponding mutants using fluorescence experiments (Figure 6F). Only the mutant E289A, with mutations at key residues of the active site (Asp268, Glu289, and Lys291), had almost complete abolishment of exonuclease activity. The four-residue mutant (Glu404Ala, Lys405Ala, Met412Ala, and Arg427Ala) showed a marked decrease in exonuclease activity, which demonstrated that R354 in the dimer state is essential for its full enzymatic activity.
Discussion
Here we report the first structure of the Cas4-like protein, R354, in the MIMIVIRE system (Figure 1), which is involved in defending against virophage invasion. Nuclease activity assays supported that R354 is probably a dual nuclease (Figures 2, 6F, and S2), and the exonuclease activity was verified by a quantitative assay (Figure 6F); however, the more explicit dual mechanism of the enzyme needs more evidence. Coupled with biochemical data, our structural studies provide insights into the unique mechanisms of R354 exonuclease activity that are enhanced by the dimerized state (Figure 6F), but not the endonuclease activity, which revealed the unique mechanism of R354. Notably, the different states of R354 indicate that the mechanism of R354 activation is distinct from those of other nuclease family members.
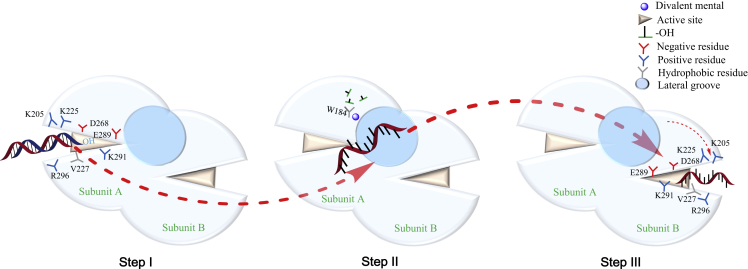
Considering that Cas4 SSO0001 (Lemak et al., 2013), Pcal 0546 (Lemak et al., 2014), and λ exonuclease (Kovall and Matthews, 1997, Zhang et al., 2011) all form toroidal oligomers, whereas R354 forms a dimer, we speculated that R354 exhibits a sequential catalytic mechanism different from those of its homologs. Indeed, the major difference between these molecules was that activated R354 dimers could be distinguished from the toroidal oligomers, suggesting that R354 possibly catalyzes one strand of dsDNA with the dimer conformation, resulting in the release of the other strand from the central channel of the subunit; the released strand then enters the other subunit for cleavage. Therefore the mechanism of R354 activation is different from those of the toroidal oligomers. A hypothetical catalytic mechanism of R354 is shown in Figure 7. The 3′-OH group of the dsDNA first binds to residues Lys205, Lys225, and Arg296, primarily via polar interactions. Moreover, hydrophobic contacts, particularly with Val227, drive the dsDNA close to the catalytic channel, and the putative dsDNA binding site was supported by ITC measurement (Figure 6E). However, the mechanisms of reaction catalysis and energy generation for the reaction are not understood. An enzyme-DNA-metal complex is formed when the dsDNA enters the active site; the divalent metal activates the scissile phosphate along with catalytic residues Asp268, Glu289, and Lys291, and residues W182 and R186 capture the leaving phosphate groups (Figures 6C–6F). One strand of dsDNA is then cleaved, whereas the other strand likely passes through the tunnel and remains bound to the lateral open groove (Figure 1D). This strand then slides into the other tunnel for cleavage, which may explain why R354 exhibits more efficient catalytic activity than the λ exonuclease.
Figure 7.
A Working Model of R354
Cartoon depicting the working model for R354 exonuclease catalytic reaction. In step I, the 3′-OH group of the dsDNA first binds to residues Lys205, Lys225, and Arg296, primarily via polar interactions. Moreover, hydrophobic contacts, particularly with Val227, drive the dsDNA close to the catalytic channel. In step II, one strand is digested into single nucleotides and the other strand slides out from the rear portal. In step III, the sliding strand is digested by subunit B without directional priority.
In summary, we report the first structure of R354, an essential viral enzyme in the novel MIMIVIRE CRISPR-Cas-like system. The structure revealed a unique mechanism of R354 activation, which exhibits dual nuclease activity. Moreover, our findings suggest that R354 is a Cas4 homolog, and therefore it extends the list of novel MIMIVIRE spacers involved in innate immunity against virophages. In addition, this protein provides a template for the identification of new CRISPR-Cas systems in other species. However, the function of R354 similar to Cas4, the component of the CRISPR system, is still unclear due to the limitations of the current study. Future work is required to determine how R354 contributes to the MIMIVIRE system function for viral immunity.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank the staff of the BL19U1 beamline at the National Center for Protein Sciences Shanghai (NCPSS) and BL17U1 at the Shanghai Synchrotron Radiation Facility for assistance during data collection. We thank S. Fan from Tsinghua University for data collection. This work was funded by a grant to W.C. from the National Natural Science Foundation of China (Grant No.31570842) and awards from the National Young Thousand Talents Program, and the Sichuan Province Thousand Talents program in China.
Author Contributions
W.C. designed the research; C.D., M.Y., and J.W. made the constructs; C.D. purified the proteins, performed the ITC and activity assays, and grew and optimized the crystals; C.D. and Y.G. collected the data; Y.G., X.Z., and W.C. determined the structure; and W.C. wrote the manuscript with contributions from the other authors. K.Y., C.N., and S.Q. contributed to the manuscript.
Declaration of Interests
The authors declare no competing financial interests.
Published: May 25, 2018
Footnotes
Supplemental Information includes Transparent Methods, seven figures, and one table and can be found with this article online at https://doi.org/10.1016/j.isci.2018.04.001.
Data and Software Availability
The atomic coordinates and structure factors of the reported crystal structures have been deposited in the Protein Data Bank (PDB), under the following accession codes: 5YET is for wild-type R354; 5YEU is for four-point mutation R354.
Supplemental Information
References
- Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., Van J.D.O. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaia M., Benamar S., Boughalmi M., Pagnier I., Croce O., Colson P., Raoult D., La Scola B. Zamilon, a novel virophage with Mimiviridae host specificity. PLoS One. 2014;9:e94923. doi: 10.1371/journal.pone.0094923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton N.C., Perona J.J. Making the most of metal ions. Nat. Struct. Biol. 2001;8:290–293. doi: 10.1038/86149. [DOI] [PubMed] [Google Scholar]
- Hudaiberdiev S., Shmakov S., Wolf Y.I., Terns M.P., Makarova K.S., Koonin E.V. Phylogenomics of Cas4 family nucleases. BMC Evol. Biol. 2017;17:232. doi: 10.1186/s12862-017-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall R., Matthews B.W. Toroidal structure of λ-exonuclease. Science. 1997;277:1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- Lemak S., Beloglazova N., Nocek B., Skarina T., Flick R., Brown G., Popovic A., Joachimiak A., Savchenko A., Yakunin A.F. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S]-cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 2013;135:17476–17487. doi: 10.1021/ja408729b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemak S., Nocek B., Beloglazova N., Skarina T., Flick R., Brown G., Joachimiak A., Savchenko A., Yakunin A.F. The CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe-2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res. 2014;42:11144–11155. doi: 10.1093/nar/gku797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur A., Bekliz M., Chabrière E., Pontarotti P., Scola B.L., Raoult D. MIMIVIRE is a defence system in mimivirus that confers resistance to virophage. Nature. 2016;531:249–252. doi: 10.1038/nature17146. [DOI] [PubMed] [Google Scholar]
- Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagens A., Tjaden B., Hagemann A., Randau L., Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol. 2012;194:2491–2500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed K.D., Lazinski D.W., Calderwood S.B., Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Steitz J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Zhang J., Mccabe K.A., Bell C.E. Crystal structures of λ exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. USA. 2011;108:11872. doi: 10.1073/pnas.1103467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.