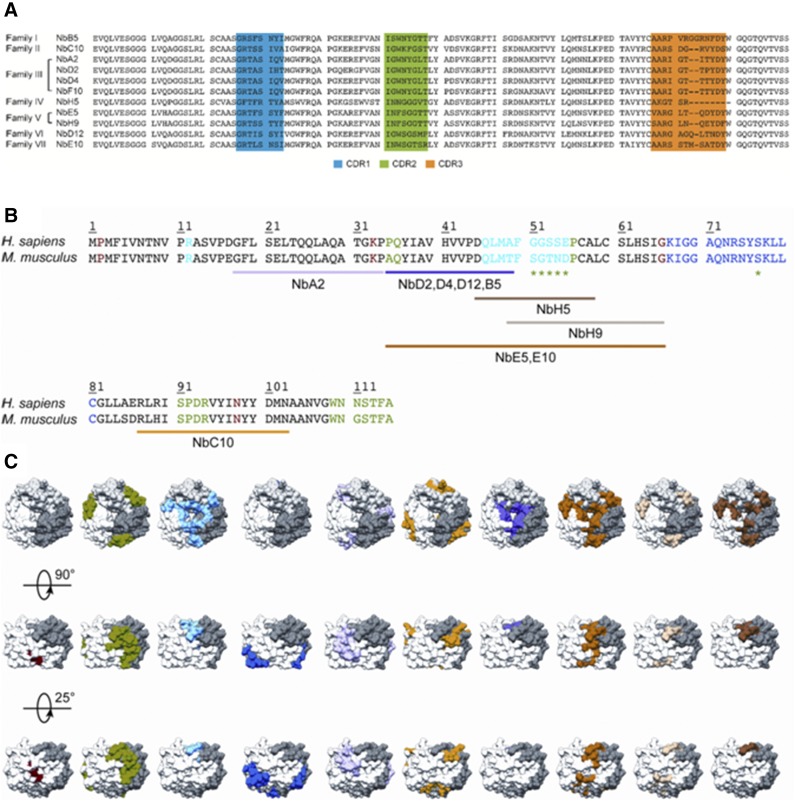
Figure 1.
Sequences of anti-MIF Nbs and their predicted binding regions on the MIF protein. A) Sequence alignment of the 11 different anti-MIF Nbs, which are organized into different families according to Kabat classification (74). Complementarity determinant region 1 (CDR1), CDR2, and CDR3 are indicated in blue, green, and orange, respectively. B) Sequence alignment of rhMIF (Homo sapiens) and rmMIF (Mus musculus). Residues involved in MIF’s tautomerase activity, CD74 binding, and the interaction with CXCR2 and CXCR4 are colored in red, green (including those indicated by the asterisk), and light and dark blue, respectively. Results of the performed epitope-mapping studies are indicated below the sequence alignment and are color coded (NbA2, pink; Nbs D2, D4, D12 and B5, purple; NbH9, light brown; Nbs E5 and E10, orange; NbC10, yellow; NbH5, dark brown; for NbF10, no definite region could be appointed). C) Results of the epitope-mapping experiment displayed on the structure of human MIF (62). MIF trimer is shown in surface representation, and each constituting monomer is shown in white, gray, and dark gray.