Figure 3.
ICLs Rapidly Induce Frequent Fork Reversal, Mediated by RAD51 and ZRANB3
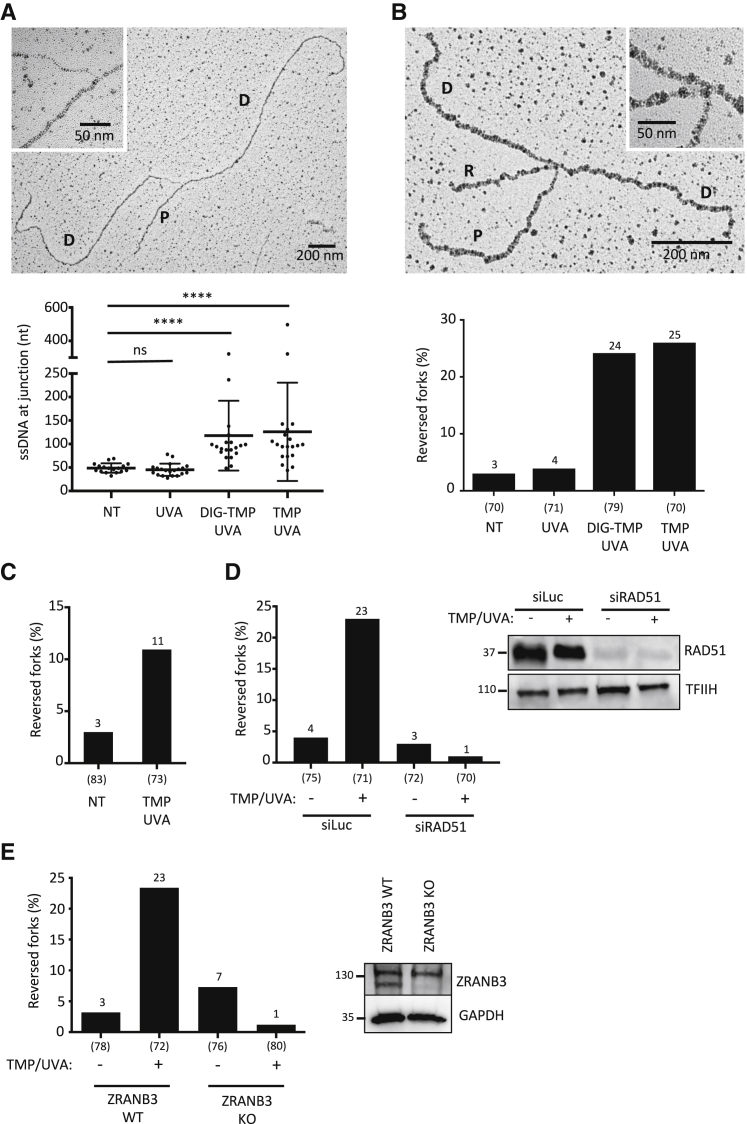
(A) Quantification of single-stranded DNA (ssDNA) at the replication junction by electron microscopy (EM). Representative image of a normal replication fork with magnification of the ssDNA region of interest (top). P, parental strand; D, daughter strand. Statistical analysis of ssDNA tract length in U2OS cells, optionally treated with either UVA alone (3 J/cm2), or DIG-TMP (5 μM) and UVA (3 J/cm2), or TMP (30 nM) and UVA (300 mJ/cm2), and collected 1 hr post-treatment for analysis (bottom). Kruskal-Wallis test (∗∗∗∗p < 0.0001; ns, not significant). Error bars indicate SD.
(B) Quantification of reversed replication forks by EM in U2OS cells treated as in (A). Representative image of a reversed fork (top) and statistical analysis (bottom). P, parental strand; D, daughter strand; R, regressed arm.
(C) Quantification of reversed forks in U2OS cells treated with TMP/UVA as in (A) but collected already 15 min post-irradiation.
(D) Percent reversed forks detected by EM in U2OS cells transfected with siLuc (control) or siRAD51 and treated optionally with TMP /UVA as in (A) (left). Western blot showing the downregulation of RAD51 protein levels (right). TFIIH, loading control.
(E) Percentage of reversed forks in ZRANB3-proficient (ZRANB3 wild-type [WT]) and ZRANB3-deficient (ZRANB3 KO) U2OS cells, which were optionally treated with TMP /UVA as in (A) (left). Western blot showing the absence of the ZRANB3 protein in ZRANB3-KO U2OS cells (right). GAPDH, loading control.
(A–E) A minimum of 70 forks were analyzed in two independent experiments. See also Table S1.