Figure 6.
Visualization of ICL Traverse as Postreplicative Sister Chromatid Junctions, Resolved by ICL Incision
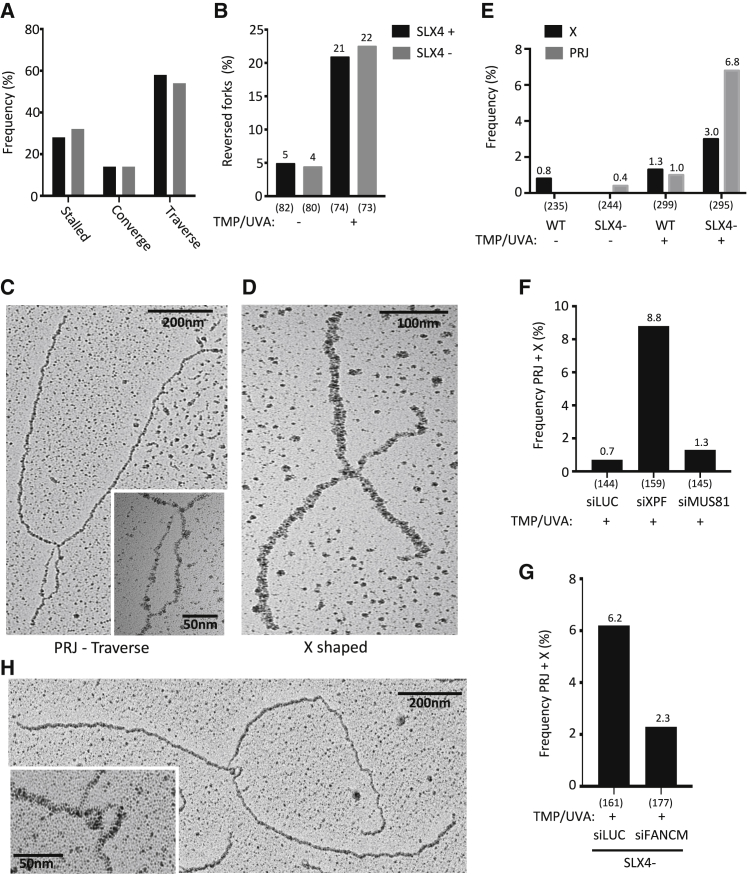
(A) Frequency of local replication patterns (as in Figure 5A) in FIT-inducible SLX4 proficient (SLX4+) or deficient (SLX4−) cells (as in Figure 4E). A minimum of 100 forks was analyzed per experiment.
(B and E) Frequency of local replication patterns determined by EM, such as reversed forks (B), postreplicative junctions (PRJs) and X-shaped molecules (E) in SLX4+ and SLX4− cells that were either left untreated or treated with TMP/UVA (30 nM/300 mJ/cm2). A minimum of 70 forks was analyzed per sample in two independent experiments.
(C and D) Representative electron micrograph showing a postreplicative junction behind a replication fork, indicative of ICL traverse (C) or an X-shaped molecule (D).
(F) Total frequency of postreplicative junction + X-shaped molecules from U2OS cells, transfected with siLuc, siXPF, or siMUS81 and treated with TMP/UVA (30 nM/300 mJ/cm2). A minimum of 70 forks was analyzed per sample in two independent experiments.
(G) Total frequency of postreplicative junction + X-shaped molecules in SLX4− cells (as in Figure 4E) transfected with siLuc or siFANCM and treated with TMP (30 nM) and UVA irradiated (300 mJ/cm2). A minimum of 70 forks was analyzed per sample.
(H) Representative image showing a postreplicative junction behind a replication fork, indicative of ICL traverse. ICL labeled with an antibody against DIG. See also Figures S5 and S6 and Tables S3–S6.