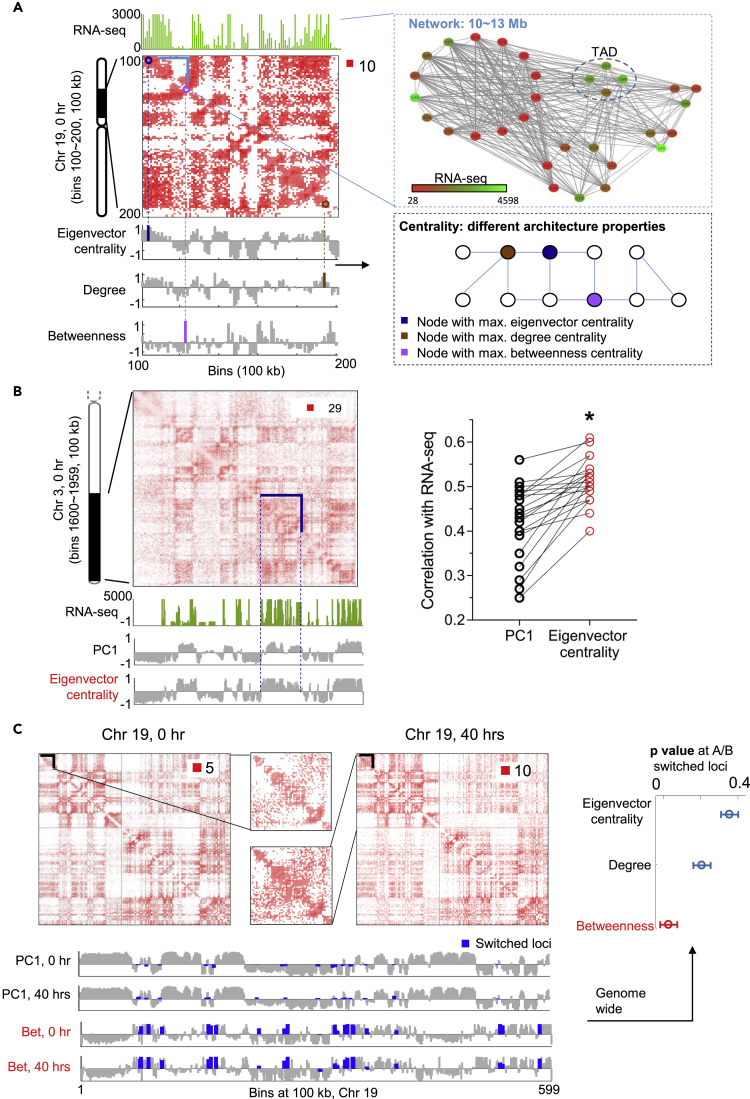
Figure 2.
Network Representation of Genomic Time Series Data
(A) Mapping genomic form (Hi-C) and function (RNA-seq) to network architecture and node dynamics. Top left: Hi-C contact map (Toeplitz normalized, Methods) and RNA-seq at 100 kb resolution for chromosome 19. Top right: Network representation in which edge width indicates the Hi-C contact number and node color implies the magnitude of RNA-seq FPKM value. Bottom left: Network features given by eigenvector centrality, degree centrality, and betweenness centrality scores. The bars marked by different colors correspond to maximum centrality values. Bottom right: An illustrative network under different centrality measures.
(B) Eigenvector centrality indicates chromatin compartments, termed A and B. Top left: Hi-C contact map of chromosome 3 at 100 kb resolution. Bottom left: RNA-seq, the first principal component (PC1) of the Hi-C correlation matrix, and eigenvector centrality (in terms of its Z score). Right: Correlation between RNA-seq, PC1, and eigenvector centrality extracted from Hi-C data for all chromosomes. Eigenvector centrality is a better indicator for chromatin compartments, marked by asterisk.
(C) Betweenness centrality indicates A/B switched loci. Top left: Hi-C contact map of chromosome 19 (100 kb resolution) at time points 0 and 40 hr. Bottom left: A/B partition and betweenness centrality (in terms of its Z score) at 0 and 40 hr. The blue color represents A/B switched bins from 0 to 40 hr. The switched loci tend to have large betweenness centrality scores. Right: Significance of betweenness centrality at A/B switched loci. The p value is determined by comparing the average betweenness value at A/B switched bins with a random background distribution of other centrality values under the same number of bins. p Values are computed for all chromosomes and shown through an error bar plot in which the circle represents the p value averaged over all chromosomes and the horizontal error bar is determined by the SD of p values for all chromosomes.