Figure 3.
Changes in Genome Architecture Precede Activation of the Myogenic Program
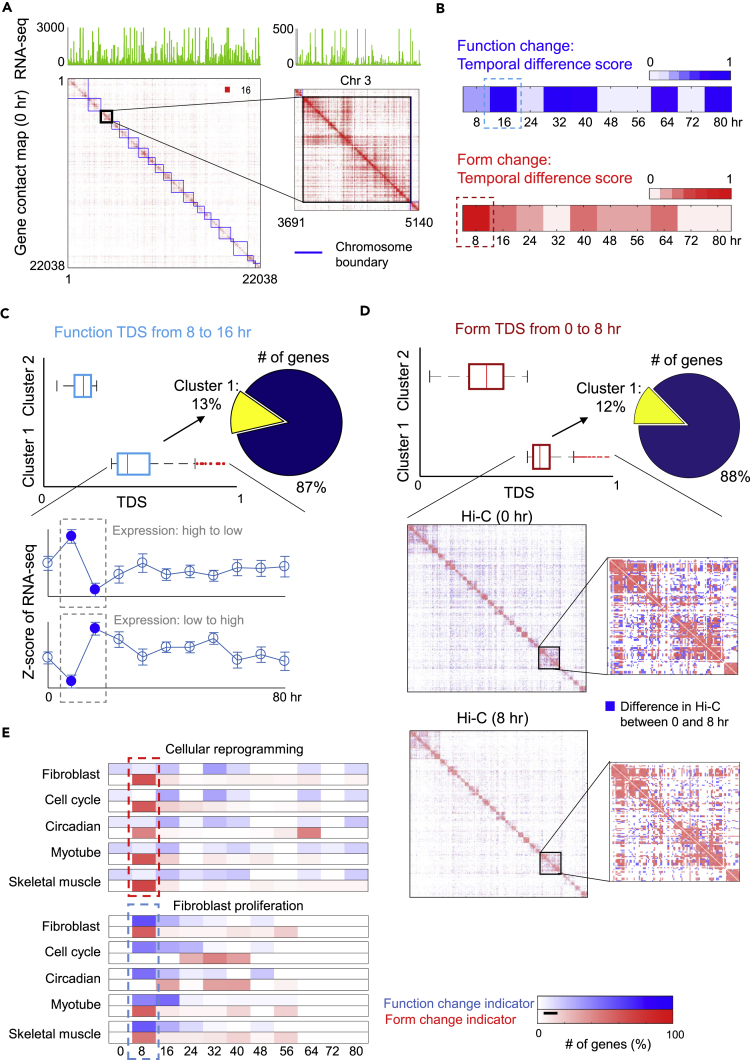
(A) Genomic architecture (form) and gene expression (function) given by a Hi-C contact map and RNA-seq. Hi-C and RNA-seq are constructed at gene-level resolution.
(B) Function and form change at successive time points evaluated by temporal difference score (TDS; Methods) of RNA-seq and network centrality features of Hi-C data, respectively. The significant form change (at 8 hr) occurs before the function change (at 16 hr).
(C) Illustration of function TDS from 8 to 16 hr. Genes are divided into 2 clusters by applying K-means to their TDS values. Cluster 1 contains genes with the largest temporal change in RNA-seq. The gene expression can either decrease or increase from 8 to 16 hr.
(D) Illustration of form TDS from 0 to 8 hr. Two gene clusters are obtained by applying K-means to their TDS values. Hi-C contact maps associated with a subset of genes in cluster 1 are shown from 0 to 8 hr, where the blue color indicates the Hi-C difference between the 2 time points.
(E) Form-function change indicators for gene modules of interest during cellular reprogramming (top) and fibroblast proliferation (bottom), respectively. Here each row represents one gene module of interest, each column represents a time step, and the amount of change, as a percentage of total change over time for each module, is depicted by color. Percentage is determined by finding the number of genes with significant form-function change for each module and time step and dividing this number by the total number of significant gene changes for each module over time (row).