Figure 5.
Increased Genomic Contacts among Myogenic Regulatory Elements Set the Stage for Reprogramming
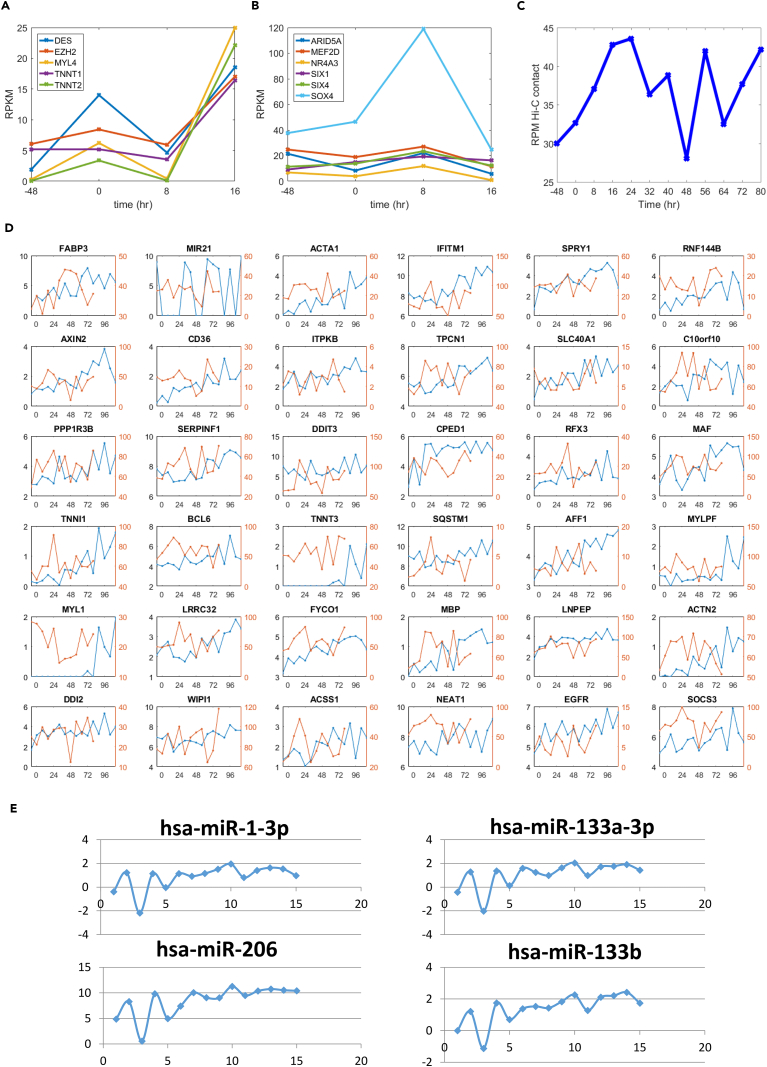
(A) Early-phase expression dynamics of genes related to muscle cell terminal differentiation and chromatin remodeling. Genes encoding proteins involved in adult muscle function, including components of the contractile apparatus (DES, MYL4, TNNT1, TNN2), and EZH2, a repressor that is involved in myogenesis.
(B) Chromatin remodeling factors and master transcription factors act cooperatively with MYOD1 to drive proliferating human fibroblasts into muscle cells. These factors include ARID5A, part of the BAF47 muscle remodeling complex that acts in cooperation with MYOD1; MEF2D, which drives differentiation of myotubes to skeletal and cardiac muscle; NR4A3 (aka NOR1) involved in differentiation of myotubes into smooth muscle; and SIX1, SIX4, and SOX4, which control the differentiation of myotubes into muscle cells.
(C) Form and function of super enhancers and associated genes over time. Average Hi-C (read per million; RPM) contact between potential super enhancer and associated gene TSS regions over time, as defined by Hnisz et al. (2013).
(D) Top upregulated SE-P genes, log2(FPKM) (blue), and SE-P Hi-C normalized contact (red; see Methods) over time.
(E) Four muscle-specific miRNAs have significantly increased expression levels in the later time points relative to the baseline control. X axis, sampling time points; y axis, log-scale differences at other time points compared with baseline (−48 hr).