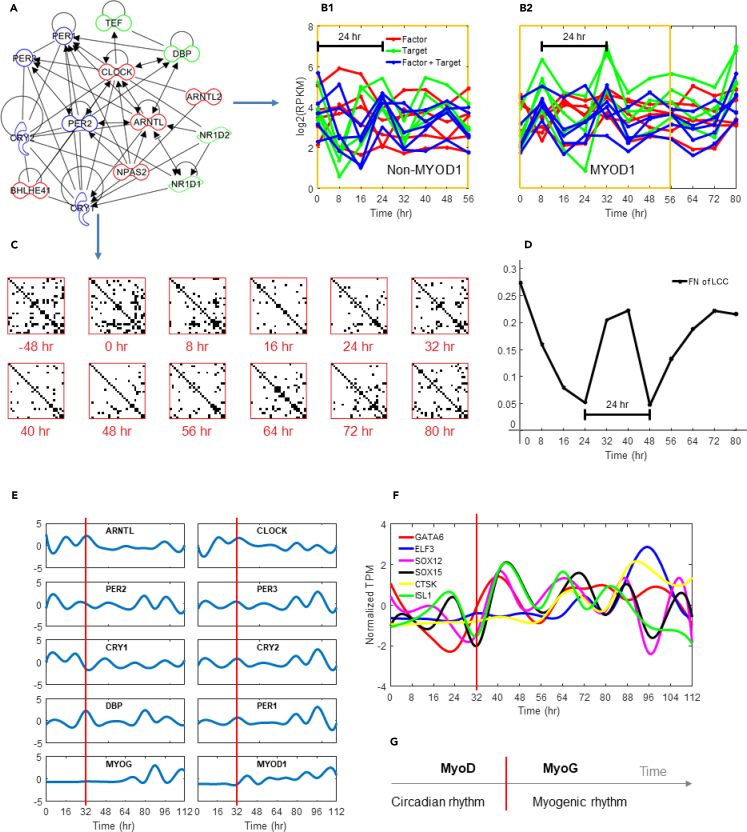
Figure 6.
Myogenic Genes Participate in Entrainment of Biological Rhythms
(A) Gene network interactions between circadian E-box genes, derived from Ingenuity Pathway Analysis.
(B) Core circadian gene expression over time. (B1) Dexamethasone synchronization. (B2) L-MYOD1 synchronization. Target and factor correspond to genes with E-box targets and TFs that bind to E-box genes, respectively.
(C) Hi-C contacts between 26 core circadian genes over time (see Table S3). Rows and columns correspond to core circadian genes; contacts are binary (i.e., any contact between genes at a given time are shown).
(D) Network connectivity of the largest connected component (LCC; Methods) of the studied Hi-C contact maps at different time points.
(E) Normalized gene expression (FPKM, cubic spline) highlighting oscillation dampening after the bifurcation time 32 hr (red line) and the switch to differentiation medium for select core circadian genes; MYOD1 and MYOG also shown.
(F) Normalized transcripts per million (TPM) of transcription factors that are targeted by MYOG or MYOD1 (ELF3) and that only showed oscillation after the critical transition at 32 hr (red line).
(G) Conceptual diagram of biological rhythm entrainment during MYOD1-mediated reprogramming, where the red line signifies the bifurcation event.