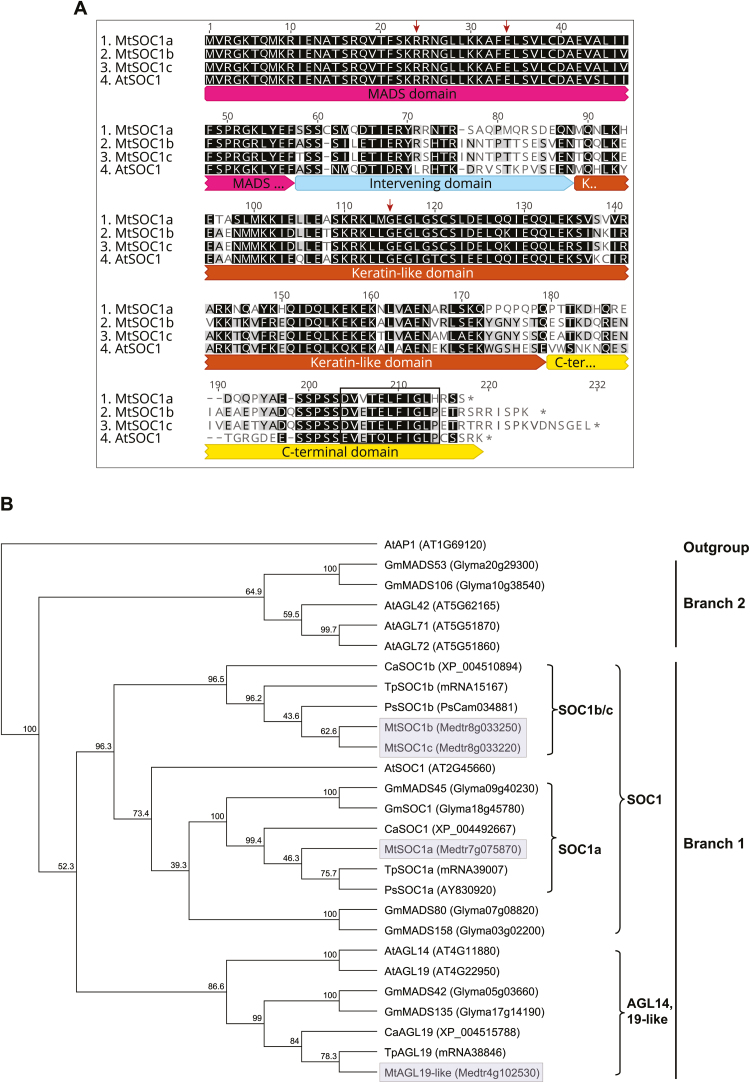
Fig. 1.
Alignment and phylogenetic analysis of MtSOC1-like proteins. (A) Protein sequence alignment of AtSOC1, MtSOC1a, MtSOC1b, and MtSOC1c indicating the conserved MADS (M), intervening (I), and Keratin-like (K) domains of the MIKC MADS box proteins. The SOC1 motif (EVETQLFIGLP) (Ding et al., 2013) is boxed and the three functional residues identified by previous functional analysis of AtSOC1 (Wang et al., 2015) are marked with arrows. Identical and similar residues are highlighted in black. (B) A consensus phylogenetic tree based on the MIK domains of TM3 members from Arabidopsis, soybean, and the temperate legumes, Medicago, pea, chick pea, and red clover rooted on Arabidopsis APETALA1 (AtAP1). The tree was generated using the Neighbor–Joining (NJ) method via bootstrap resampling with a support threshold of 39%. The numbers indicate the bootstrap values based on 1000 replicates. The branch numbering is from Dorca-Fornell et al. (2011). The four Medicago proteins are highlighted in grey. At, Arabidopsis thaliana; Ca, Cicer arietinum (chick pea); Gm, Glycine max (soybean); Mt, Medicago truncatula; Ps, Pisum sativum (pea); Tp, Trifolium pratense (red clover).