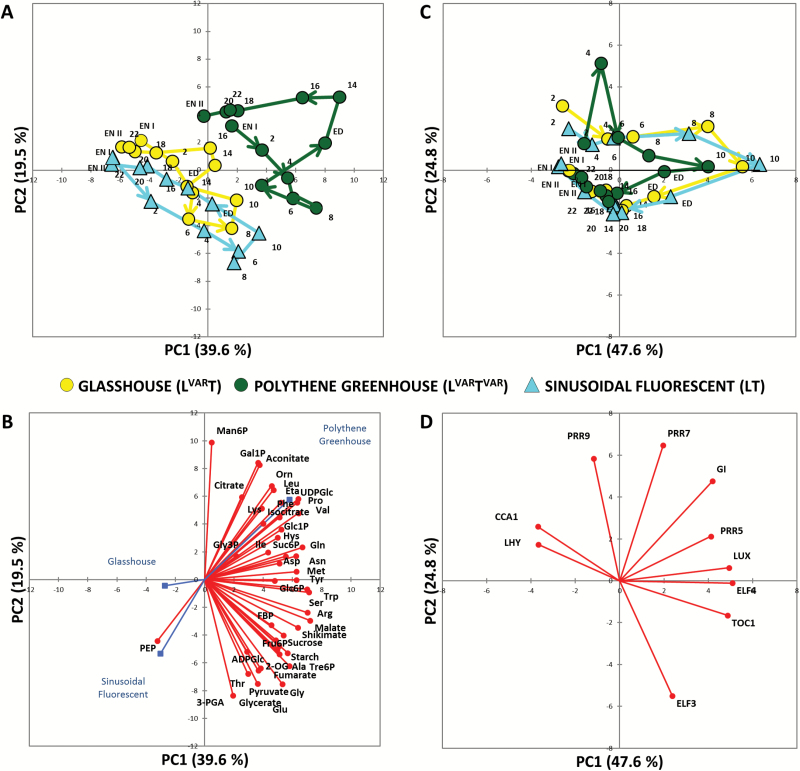
Fig. 1.
Principal component analysis (PCA) of metabolite and core circadian clock gene data from Arabidopsis plants. PCA of metabolite data (A) and of core clock genes (C) from plants grown around the vernal equinox in 2015 in a naturally illuminated and temperature-controlled glasshouse (LVART, yellow circles) or a naturally illuminated polythene greenhouse with a less controlled temperature (LVARTVAR, green circles). Plants were also grown in a controlled-environment chamber with a 12 h photoperiod and daily light integral (DLI) of 12 mol m−2 d−1. The artificial illumination was provided by white fluorescent tubes with a sinusoidal (LT, cyan triangles) light profile during the day. Numbers indicate the time of harvest in hours after dawn (Zeitgeber time, ZT); ED, end of day (ZT12); EN I, end of preceding night (ZT0); EN II, end of night (ZT24); and the diurnal trajectories are indicated by arrows. The percentages of total variance represented by principal component 1 (PC1) and principal component 2 (PC2) are shown in parentheses. (B and D) The loadings of individual metabolites or genes in PC1 and PC2. FBP, fructose 1,6-bisphosphate; Eta, ethanolamine; Orn, ornithine.