Figure EV1. m5C38‐tRNAA sp GUC dependency on Q in HeLa cell culture and tRNA level quantifications.
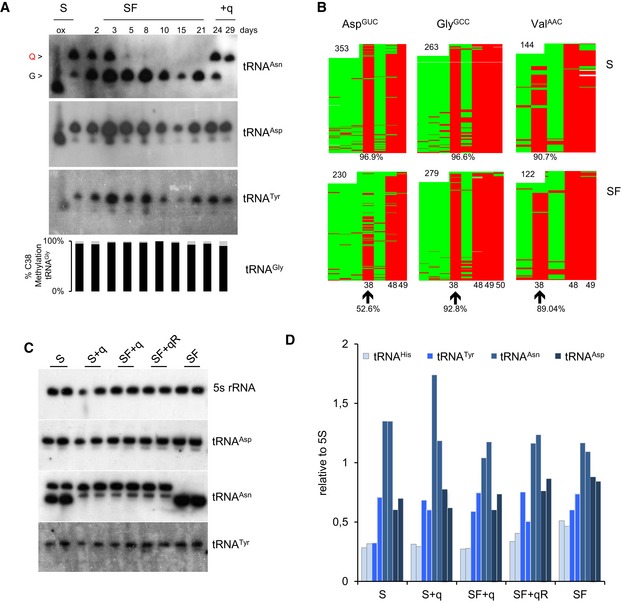
- APB Northern blot using tRNAAsn probe shows that SF medium depletes Q‐tRNA. No separate Q‐ and G‐tRNAAsp and Tyr bands were detected. m5C38 levels are measured by 454 bisulfite sequencing. A concomitant robust reduction in Q‐tRNAHis and m5C38‐tRNAAsp GUC is observed in HeLa cells cultivated in SF medium. Both queuosinylation and methylation levels could be restored by the addition of queuine to the SF medium for 3 or 8 days after 21 days in SF medium. The differential migration is eliminated by oxidizing the ribose with periodate, producing a single faster migrating band (ox).
- Bisulfite sequencing maps from a biological replicate culture under S and SF for 3 weeks are depicted. Each row represents one sequence read and each column a cytosine residue. Green boxes represent unmethylated cytosine residues, and red boxes indicate methylated cytosine residues. Sequencing gaps are shown in white. Numbers in the maps indicate the number of reads. The position of specific cytosine residues and level of C38 methylation are indicated at the bottom.
- APB Northern blot analysis that complete Fig 2B with tRNAAsp, tRNAAsn, tRNATyr, and 5S rRNA as a loading control.
- Quantitative analysis of signals for tRNAs at the indicated culture conditions. tRNA signal intensities were normalized to 5S rRNA levels for each of the two replicates.
