Figure 3. Translocase activity of Irc5 protects against DNA damage accumulation and enables replication completion.
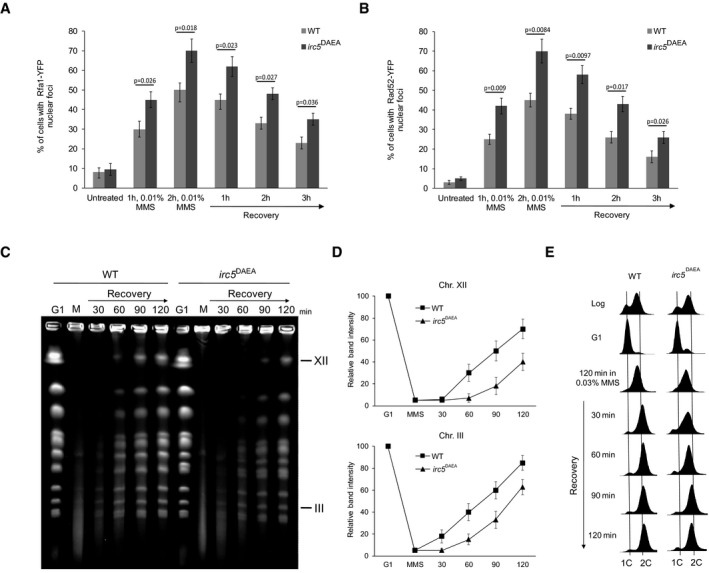
- Lack of Irc5 translocase activity results in elevated number of cells with Rfa1‐YFP foci. At indicated time points, samples of wild‐type (TB042) and irc5 DAEA (TB043) cells were collected, washed, and processed for microscopic analysis. Error bars represent mean value ± standard deviations of mean (n = 5). Unpaired t‐test was used to calculate the P‐value.
- Increased levels of bright Rad52‐YFP foci in cells expressing irc5 DAEA lacking ATPase activity. At indicated time points, samples of wild‐type (TB044) and irc5 DAEA (TB045) cells were collected, washed, and processed for microscopic analysis. Error bars represent mean value ± standard deviations of mean (n = 5). Unpaired t‐test was used to calculate the P‐value.
- Translocase activity of Irc5 is important for completion of DNA replication after MMS treatment. PFGE was performed as in Fig 2B using wild‐type (EMD09) and irc5 DAEA (EMD10) strains.
- Quantification of data presented in (C). Error bars represent mean value ± standard deviations of mean for the intensity values of three distinct chromosome bands representing chromosome XII or III (n = 3). Values for G1 were set to 100.
- Samples used for PFGE in (C) were processed for FACS analysis.
Source data are available online for this figure.
