Figure EV3. Generation and functional characterization of Charme −/− mice.
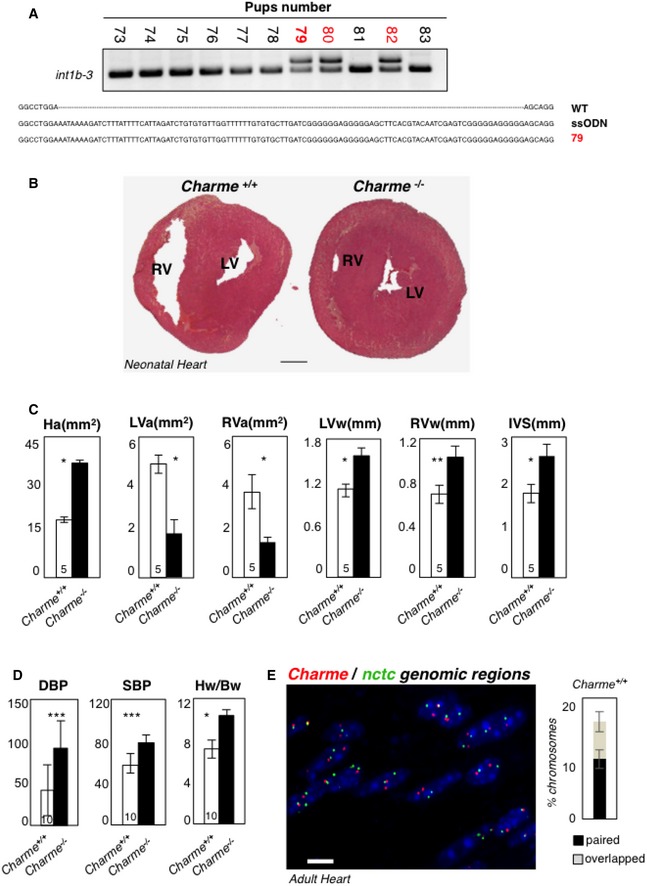
- DNA extracted from F0‐generation mice was analyzed by PCR amplification for genotyping. The position of PCR primers is indicated in Fig 1A. Electrophoretic analyses of PCR products and multiple sequence alignment (performed by Muscle‐3.8) indicated the insertion of the entire poly(A)/2×MAZ cassette on a single allele in the #79, #80, and #82 pups (red). The #79 was selected as founder for successive breeding.
- Hematoxylin and eosin staining of transverse sections of the myocardium in 2‐day‐old Charme +/+ (left) and Charme −/− (right) mice. Left (LV) and right (RV) ventricles are indicated. Scale bar = 0.5 mm.
- Morphometric analyses of heart area (Ha), left and right ventricle areas (LVa and RVa), left and right ventricle walls (LVw and RVw), and interventricular septum (IVS) in Charme +/+ and Charme −/− mice.
- Systolic (SBP) and diastolic (DBP) blood pressures and heart/body weight ratio (Hw/Bw) in Charme +/+ and Charme −/− mice. Values represent mean ± SEM of replicates. The numbers of mice tested for each group are indicated in the white bars. Mean ± SD ND values are shown. **P < 0.01, ***P < 0.001, unpaired Student's t‐test.
- DNA/DNA FISH in adult cardiac tissues. Charme +/+ adult cardiac tissues exhibit spatial proximity of Charme and nctc genomic regions. The percentages of chromosome 7 showing paired and overlapped signals are 12% and 7%, respectively, in a total of 412 nuclei analyzed. Scale bar = 10 μm.
Source data are available online for this figure.
