-
A
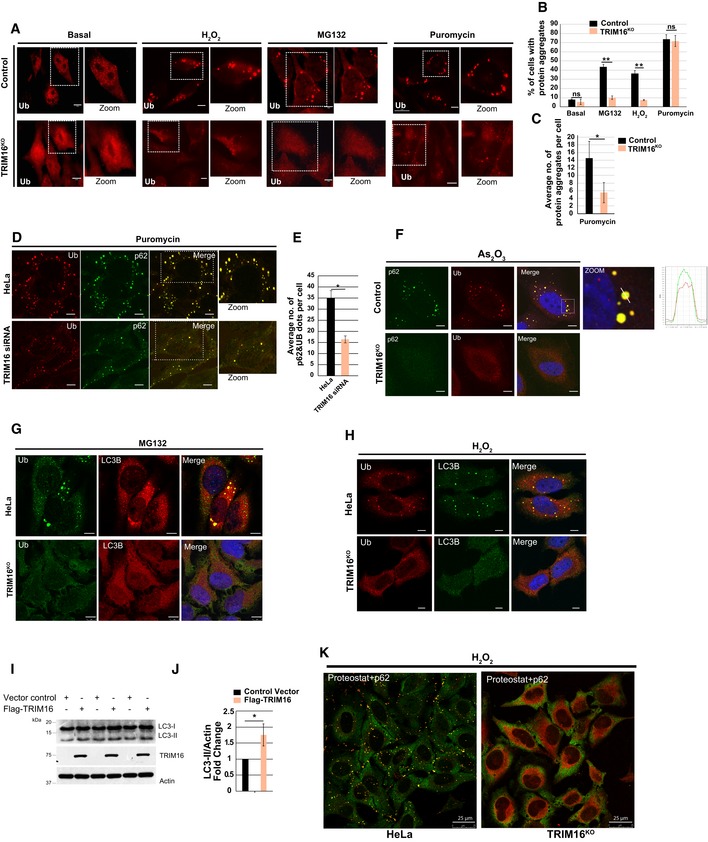
Confocal images of HeLa and TRIM16KO cells, untreated or treated with MG132 (20 μM, 2 h), H2O2 (200 μM, 2 h), and puromycin (5 μg/ml, 2 h), and processed for immunofluorescence (IF) analysis with ubiquitin (FK2) antibody.
-
B
HeLa and TRIM16KO cells were visualized, and cells with more than two visible aggresomes/ALIS were considered positive. Mean ± SD, n = 3, **P < 0.005 (Student's unpaired t‐test).
-
C
Total numbers of visible Ub puncta in each cell were counted and plotted. Data are from ≥ 10 microscopic fields for each condition, n = 3, mean ± SD, *P < 0.05 (Student's unpaired t‐test).
-
D
Representative confocal images of control siRNA and TRIM16 siRNA transfected cells treated with puromycin (5 μg/ml, 2 h), where IF analysis was conducted with Ub and p62 antibodies.
-
E
The graph shows the average number of Ub‐p62 co‐localized aggregates per cell from the experiment in panel (D). Mean ± SD, n = 3, *P < 0.05 (Student's unpaired t‐test).
-
F
Representative confocal images of HeLa and TRIM16KO cells treated with As2O3 (2.5 μM, 2 h), where IF analysis was conducted with Ub and p62 antibodies. Right panel: Fluorescence intensity line is tracing corresponding to a white line in zoom panel.
-
G, H
Representative confocal images of HeLa and TRIM16KO cells treated with (G) MG132 (20 μM, 2 h) or (H) H2O2 (200 μM, 2 h) and processed for IF analysis with Ub and LC3B antibody.
-
I
WB analysis of LC3B levels in lysates of cells transfected with control vector or Flag‐TRIM16.
-
J
Densitometric analysis of WB normalized with actin. Mean ± SD, *P < 0.05 (Student's unpaired t‐test), n = 3.
-
K
Representative confocal images HeLa and TRIM16KO cells treated with H2O2 (200 μM, 2 h) and processed for IF analysis with ProteoStat dye and p62 antibody.
Data information: Unless otherwise stated, scale bar: 10 μm.