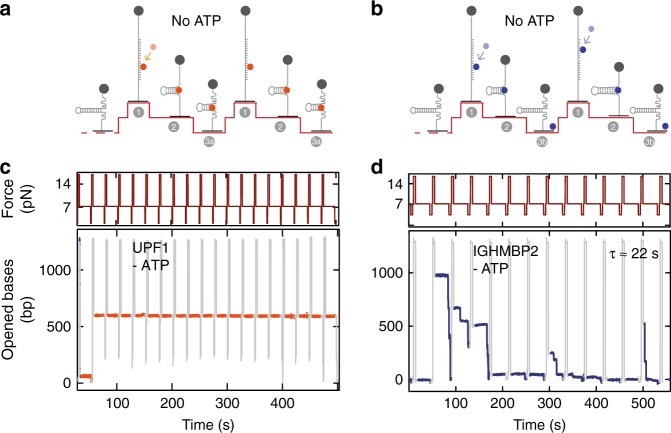
Fig. 2.
Single-molecule binding assay (SMBA) to assess helicase binding. a, b Schematic representation of the single-molecule binding assay cycle in absence of ATP. DNA hairpin is initially completely closed. Step 1, a force >15 pN is applied to fully unzip the DNA hairpin which is kept in an open state for 4 s to allow a helicase in solution to bind to the ssDNA. Step 2, force reduction to 7 pN refolds hairpin to its natural state, unless a helicase is bound. Force is held at 7 pN for 20 s leading to helicase residence on substrate over several cycles (3a) or expulsion after one or a few cycles (3b). Step 3, force is reduced <3 pN to check that the hairpin can still completely close. The whole process is repeated several times. c Upf1 remains bound to the substrate at its initial position and blocks hairpin closure over all force cycles. d IGHMBP2 transiently binds then falls off the substrate after one or a few cycles. Values on y-axis in c and d indicate hairpin opening during step 2 of the binding assay described in a and b