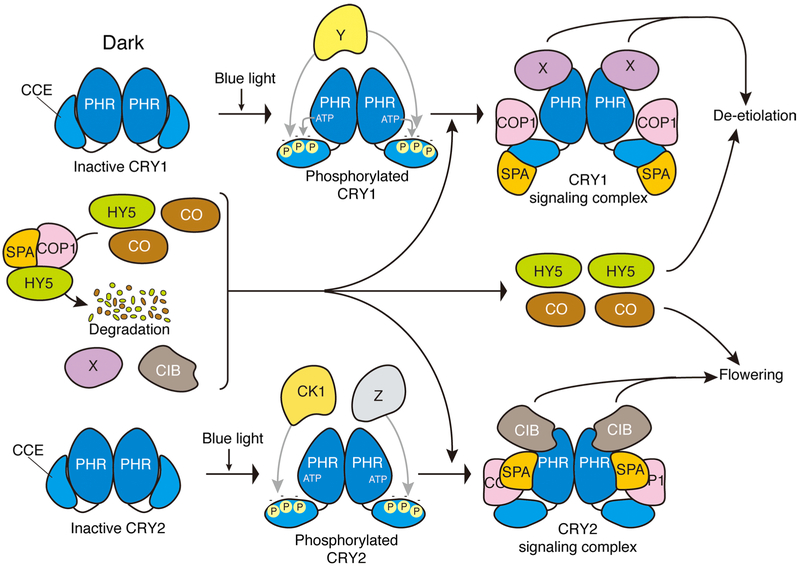
Fig. 1.
A model depicting cryptochrome signal transduction mechanisms in Arabidopsis. Cryptochromes (CRY) are composed of two domains, the PHR domain and the CCE domain, and form homodimers through the PHR domains. Upon blue light activation, CRYs undergo conformational changes in both the PHR and CCE domains to transduce signals to downstream components. Light-dependent phosphorylation at multiple sites of the CCE domain produce negative charges (−) at these sites via autophosphorylation and possibly by unknown kinases (Y) in the case of CRY1, and by CK1 and unknown kinases (Z) in the case of CRY2. The negative charges result in the electrostatic repulsion of CCE domain from PHR domain. In the dark, COP1 and SPA constitute an active complex to promote E3 ubiquitin ligase activity, leading the degradation of transcription factors such as HY5 and CO, positive regulaters for de-etiolation and flowering, respectively. In the light, photoactivated CRY1 constitutes the CRY1 signaling complex, in which CRY1 dissociates COP1 from SPA by direct binding via the CRY1 CCE domain to inactivate COP1/SPA complex. This results in the accumulation of HY5 and probably CO, and CRY1 binds with unknown factor (X) via the CRY1 PHR domain to activate the COP1 independent pathway, to promote primarily de-etiolation. Photoactivated CRY2 directly binds with SPA via the PHR domain to inactivate COP1/SPA complex by tightening the connection between COP1 and CRY2. CRY2 also binds with CIBs through the PHR domain to promote flowering. CK1 Casein kinase1, CCE cryptochrome C-terminal extension, CIB CRYinteracting basic-helix-loop-helix, CO CONSTANS, COP1 CONSTITUTIVE PHOTOMORPHOGENIC 1, FT FLOWERING LOCUS T, HY5 LONG HYPOCOTYL5, PHR photolyase-homologous region, SPA SUPPRESSOR OF PHYA-105