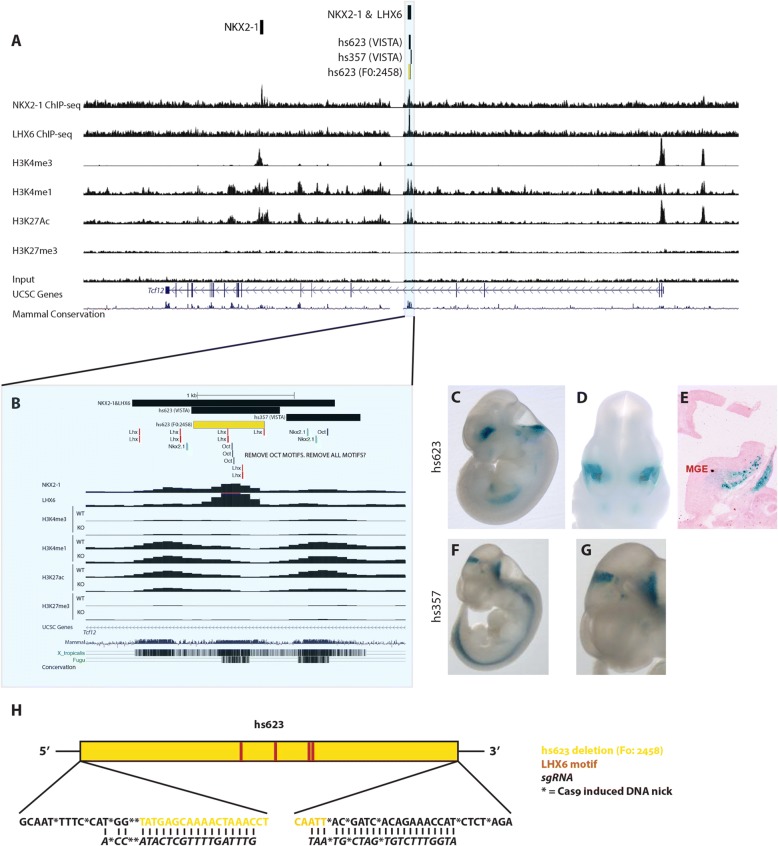
Fig. 2.
Deletion of cis-regulatory element hs623 in vivo. a Genomic region of the Tcf12 locus with the ChIP-seq datasets and genomic features shown; NKX2–1 ChIP-seq, LHX6 ChIP-seq, H3K4me3, H3K4me1, H3K27ac, H3K27me3, UCSC genes and mammalian conservation. Histone 3 modifications in MGE at E13.5. Hs623 region framed and highlighted in blue. b Higher resolution view of the hs623 region with the same ChIP-seq datasets as in Fig. 2a. Called NKX2–1 & LHX6 binding region, VISTA regions, deleted hs623 region (yellow) and NKX2–1, LHX6 and OCT consensus motifs labeled at the top of the browser. c-g VISTA database transgenic embryos showing in vivo activity of hs623 and hs357 at E11.5. h Schematic description of the generated hs623 deletions (5 founders). The distribution of LHX6 consensus motifs in hs623 are indicated. Founder 2458 was used for the analysis presented in this paper