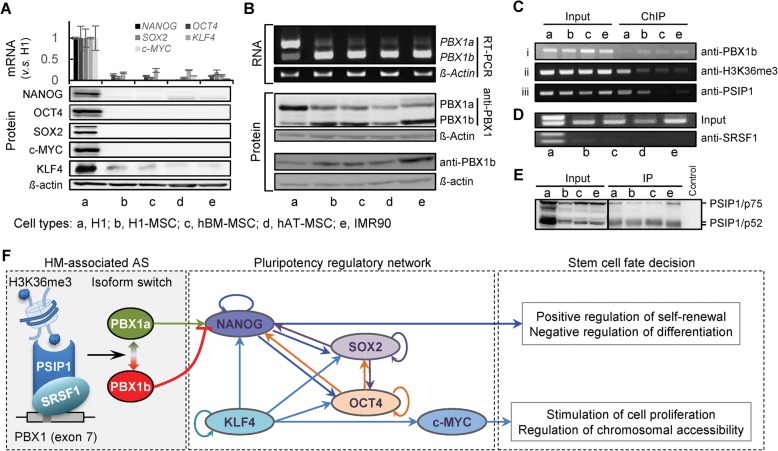
Fig. 6.
Isoform switch of PBX1 links H3K36me3 to hESC fate decision. a qRT-PCR and western blot show the expression levels of Yamanaka factors in H1, MSC, and IMR90 cells. Whiskers denote the standard deviations of three replicates. b RT-PCR and western blot show the isoform switches between PBX1a and PBX1b from H1 cells to differentiated cells. c i. ChIP-PCR shows the differential binding of PBX1b to NANOG promoter in H1 cells and differentiated cells; ii. ChIP-PCR shows the reduced H3K36me3 signal in differentiated cells; iii. ChIP-PCR shows the differential recruitment of PSIP1 to exon 7 of PBX1. d RIP-PCR show the differential recruitment of SRSF1 around exon 7 of PBX1. e Co-IP shows the overall physical interaction between PSIP1 and SRSF1 in all studied cell types. f The mechanism by which H3K36me3 is linked to cell fate decision by regulating the isoform switch of PBX1, which functions upstream of the pluripotency regulatory network. Also see Additional file 1: Figures S9, S10