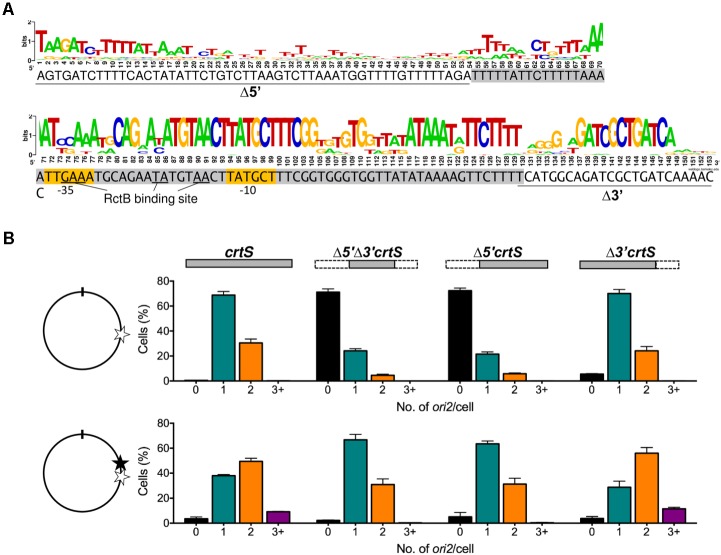
FIGURE 1.
The 5′ but not the 3′ end of crtS is critical for its function. (A) The 153 bp crtS sequence from V. cholerae overlaid with a WebLogo (Crooks et al., 2004), generated from different Vibrio species showing varied conservation along the length of the sequence, as in Kemter et al. (2018). Gray highlighted sequence denotes the minimal 70-bp that retain copy-number enhancement function of crtS in E. coli. The flanking 5′ and 3′ sequences studied here are underlined. Yellow highlighted sequences denote the predicted –35 and –10 elements of the sigma-70 promoter, PcrtS. (B) The effect of crtS and its truncated derivatives on Chr2 replication is shown by histograms of ori2 foci numbers per cell in V. cholerae strains where crtS was replaced at its native locus with the truncated derivatives (top row), and where the same mutant strains had, at 10 kb upstream, a second full length crtS copy (bottom row). On the left of the histogram is shown the approximate location of crtS copies in Chr1, where the native locus is indicated by an empty star and the locus with the added crtS copy by a filled star. The position of the ori1 is denoted by a tick-mark. The strains used were: intact crtS (CVC3058, top; CVC3061, bottom), Δ5′Δ3′crtS (CVC3228, top; CVC3247, bottom), Δ5′crtS (CVC3227, top; CVC3246, bottom), Δ3′crtS (CVC3226, top; CVC3245, bottom). Note that deletion of the upstream 53 bases (Δ5′) severely compromises replication-triggering function of crtS as evidenced by the appearance of cells with zero ori2 foci. Data represent mean ± SEM (standard error of mean) of at least 1000 cells imaged from three biological replicates.