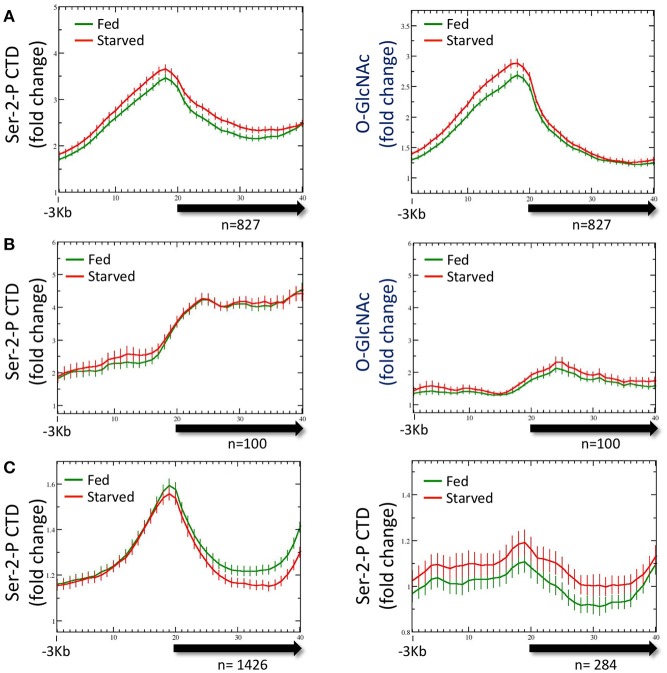
Figure 3.
RNA Polymerase II profiles for O-GlcNAc marked genes are similar in both starved and fed conditions. (A) RNA Pol II Ser-2-P (ab5095), representative of multiple Pol II antibodies used, and O-GlcNAc ChIP profiles for the 827 O-GlcNAc marked gene set with axis and gene model displays as described in Figure 1; the O-GlcNAc profile is duplicated from Figure 1A (right panel) for side by side comparison to the corresponding Pol II data. Very little difference was observed for the Pol II distribution in comparing fed and starved conditions. Note that while Pol II and O-GlcNAc profiles are similar in the promoter regions, the Pol II signal extends into the gene body whereas O-GlcNAc does not. (B) To determine if the O-GlcNAc marked gene set behavior was unusual in being unchanged regardless of feeding condition, we examine several gene sets that were selected using different criteria. Show is a set of 100 genes that had high levels of RNA Pol II within the gene body, but low promoter region O-GlcNAc signals. This gene set also shows nearly identical profiles for Pol II (left panel) in either starved or fed conditions, lacks a strong signal for promoter region O-GlcNAc (right panel), and has an overall profile that is quite distinct from the 827 O-GlcNAc marked genes. (C) Genes with nutritionally responsive expression have corresponding changes in Pol II occupancy. As many gene sets showed similar profiles for Pol II in both starved and fed conditions, we identified genes that were nutritionally responsive to these two conditions based only on changes in gene expression (log2 fold change greater than or equal to 1.5 when comparing starved and fed values). The left panel shows the Pol II profile, represented by the Ser-2-P data, for 1426 genes that are up-regulated in fed conditions compared to starved values. As expected, the fed Pol II profile increases in the gene body of these genes. For genes that are up-regulated in starved conditions (n = 284; right panel), the starved Pol II profile is greater than the fed. Thus, the Pol II ChIP profile changes are consistent with the gene expression data for those gene showing the most dynamic response to feeding condition whereas Pol II occupancy profiles for nutritionally unresponsive genes are correspondingly homeostatic.