Figure 3.
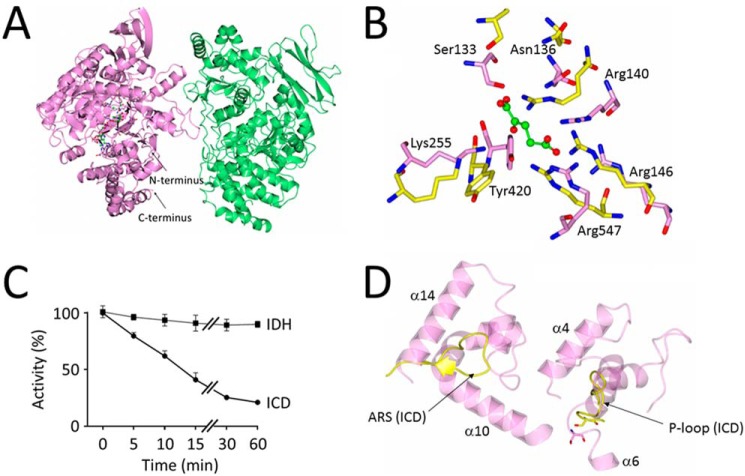
Structure and regulation of P. aeruginosa IDH. A, cartoon schematic of IDH from P. aeruginosa. Chain A (mauve) contains one molecule each of NADP+ and α-ketoglutarate bound in its active site. Chain B (light green) does not contain any bound small molecules. The two IDH chains in the asymmetric unit do not show enough interprotein contacts to warrant being labeled as protomers in a dimer. B, close-up view of the conserved active site residues in P. aeruginosa IDH (light pink, PDB code 6G3U) and P. aeruginosa ICD (yellow, PDB code 5M2E). Residue numbering is based on the IDH sequence. C, P. aeruginosa ICD, but not IDH, is inactivated by AceK-dependent phosphorylation. Shown is the loss of isocitrate dehydrogenase activity over time following treatment with AceK/ATP. Reaction mixtures (200 μl) contained 100 mm Tris-HCl (pH 7.0), 1 mm ATP, 2 mm MgCl2, 5 μg of purified P. aeruginosa AceK, and 10 μg of P. aeruginosa ICD or IDH (as indicated). Reactions were allowed to proceed at 37 °C, and at the indicated times, aliquots were withdrawn and assayed immediately for isocitrate dehydrogenase activity, as described under “Experimental procedures.” Activity was considered to be 100% at T0. D, the architecture of the active site is different in ICD and IDH. Despite the conserved constellation of active site residues (B) in P. aeruginosa ICD and IDH, the P-loop containing the phosphoserine in ICD (yellow) and the AceK recognition segment (ARS) are replaced in IDH by two helices, α10–α14 and α4–α6, respectively. This altered arrangement presumably prevents AceK from accessing the active site serine in IDH.