Figure 1.
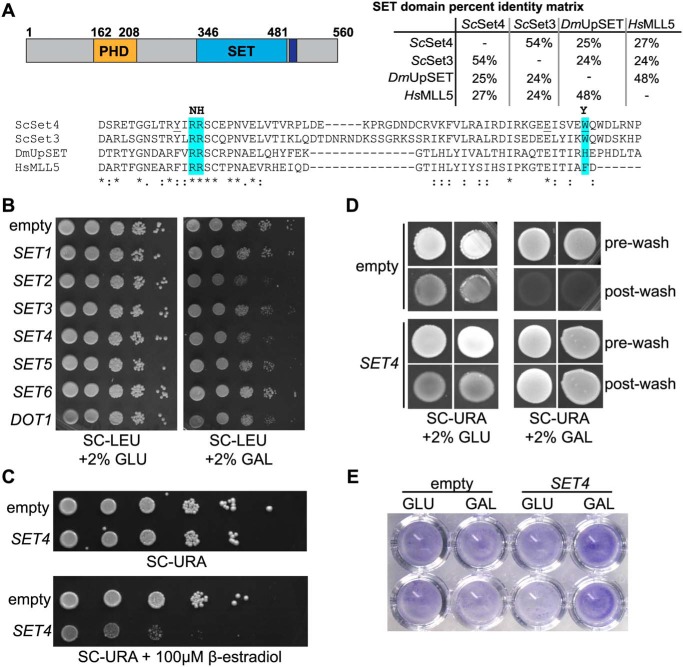
Overexpression of SET4 is deleterious and promotes cell adhesion. A, the domain structure of Set4 (left), indicating the position of the PHD finger (orange), the SET domain (blue), and the post-SET domain (purple). A percentage identity matrix (right) compares the SET domains of S. cerevisiae Set4 (ScSet4), Set3 (ScSet3), D. melanogaster UpSET (DmUpSET), and H. sapiens MLL5 (HsMLL5). Percentage identity was determined using Clustal Omega. Shown is a sequence alignment of the most conserved region of the SET domain among ScSet4, ScSet3, DmUpSET, and HsMLL5. Amino acids highlighted in blue represent differences from canonical SET domains in regions required for SAM and lysine binding (20). Amino acids predominantly found in other SET domains are indicated in boldface type at the top of the alignment. Underlined amino acids within the Set4 sequence indicate mutations used in Fig. 8C. B, WT yeast (yEG001) carrying plasmids expressing known and putative histone methyltransferases under the control of the GAL1 promoter. 10-Fold serial dilutions were spotted on either SC-LEU with 2% glucose (GLU) or 2% galactose (GAL) and grown at 30 °C for 3 days. C, strains harboring a β-estradiol–inducible artificial transcription factor and either the empty pMN3 vector or pMN3-SET4 were grown in SC-URA, and 10-fold serial dilutions were spotted on SC-URA or SC-URA with 100 μm β-estradiol and grown at 30 °C for 3 days. D, the Σ1278b strain with pRS316-GAL1p-SET4 or an empty vector was grown on SC-URA with either 2% glucose or 2% galactose for 10 days to promote adhesion to the agar and invasion. Images were obtained before washing (pre-wash) and after gentle washing with water to remove nonadherent cells (post-wash). E, crystal violet staining of Σ1278b cells containing an empty vector or pRS316-GAL1p-SET4 grown in either 2% glucose (GLU) or 2% galactose (GAL) and adhered to 96-well polystyrene plates, as described (46).