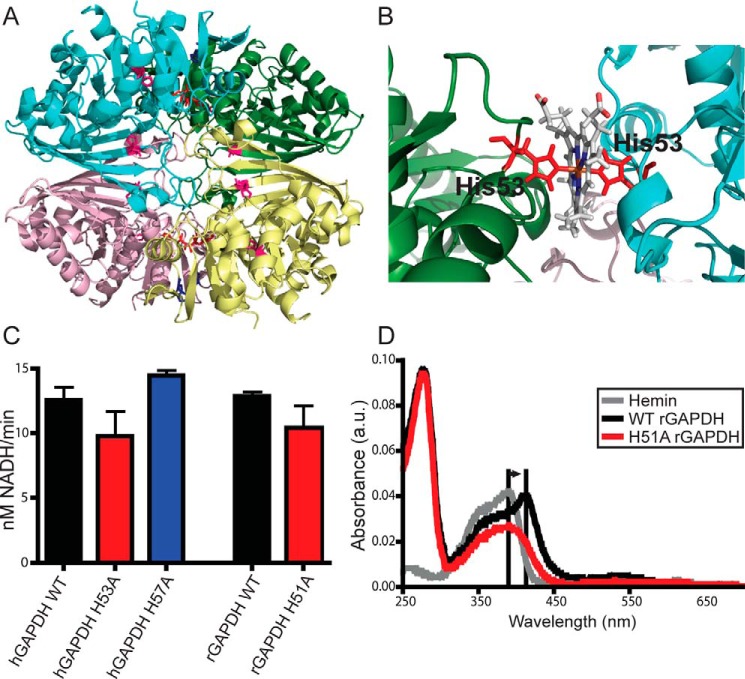
Figure 2.
GAPDH heme binding involves a specific His residue. A, human GAPDH tetramer from PDB code 1znq. Conserved histidines are shown as sticks; His-53 is in red; His-57 is in blue, and His-111 and His-179 are in magenta. B, close-up of subunit interface showing a representative structure from molecular dynamics simulations of the modeled human GAPDH–heme complex. C, glycolytic activity of GAPDH WT and mutants measured using conversion of NAD+ to NADH. Values represent mean ± S.E. (n = 5, no significance). D, UV-visible spectra of rabbit GAPDH in the presence of heme. Free hemin displays a Soret peak at 390 nm. Histidine ligation of the heme shifts this peak to 415 nm. WT but not H51A (His-53 equivalent) rGAPDH displays this spectral shift indicative of heme binding through histidine ligation. Changes to the heme absorbance trace in the presence of H51A rGAPDH indicates protein–heme binding without histidine ligation.