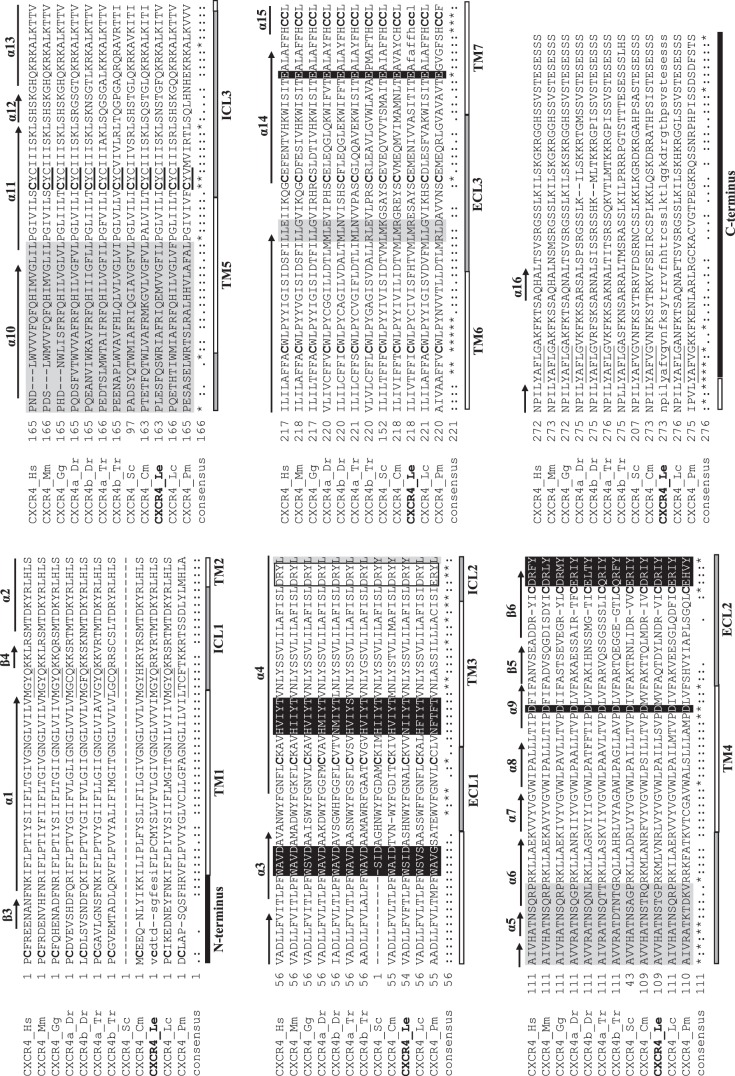
Fig. 2.
Annotated multiple alignment of vertebrate chemokine receptor type 4 (CXCR4; subset of the vertebrate chemokine ligand multiple alignment; see Supplemental Fig. S4). The little skate sequence (CXCR4_Le) is in boldface. The portion of the little skate sequence retrieved from sequencing is written in capital letters, whereas the inferred portion is written in lowercase letters. The characteristic C-X-C motif is denoted by a boldfaced underline. Sequences involved in chemokine binding are highlighted in black with white letters, whereas sequences involved in dimerization are highlighted in light gray. The boxed sequence (DRY for all species except lamprey) denotes an area that is critical for signaling. Uniformly conserved cysteines are in boldface. Sequences responsible for turns in the secondary protein structure in human CXCR4 are underlined. Individual β-strands (β3–6) and α-helices (α1–16) inferred from the human CXCR4 sequence are denoted by arrows above the alignment. For the consensus sequence, identical amino acids are denoted by an asterisk (*), whereas those with low similarity (i.e., 50%) are denoted by a period (.), and those with high similarity (i.e., 70%) are denoted by a colon (:). Under the consensus sequence, the NH2 terminus and COOH terminus are shown as black bars, transmembrane domains (TM1–7) as white bars, and extracellular loops (ECL1–3) and intracellular loops (ICL1–3) as gray bars. The sequence accession numbers are described in Supplemental Table S1, and species abbreviations are as follows: Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Pm, lamprey (Petromyzon marinus); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).