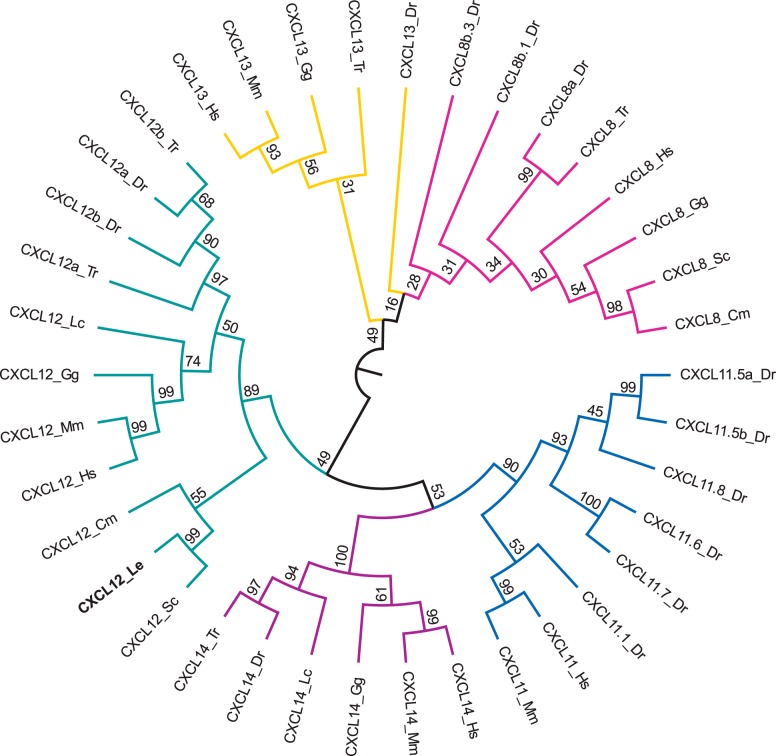
Fig. 3.
Phylogenetic tree of vertebrate chemokine ligands from the C-X-C motif chemokine ligand (CXCL) 8 (pink), CXCL11 (blue), CXCL12 (green), CXCL13 (yellow), and CXCL14 (purple) gene families. The tree was constructed in the MEGA6 program (54) using a MuscleWS multiple alignment. The evolutionary history was inferred by using the maximum likelihood method based on the LG model (30) with invariable sites (+I; 6.1275% sites). A discrete γ-distribution was used to model evolutionary rate differences among sites (+G; five categories, parameter = 3.6495). Node values represent %bootstrap confidence derived from 10,000 replicates. The little skate CXCL12 sequence (CXCL12_Le) is in boldface. The analysis included 38 amino acid sequences. All positions containing gaps or missing data were eliminated, resulting in a total of 65 positions in the final data set. The sequence accession numbers are described in Supplemental Table S1, and species abbreviations are as follows: Cm, elephant shark (Callorhincus milii); Dr, zebrafish (Danio rerio); Gg, chicken (Gallus gallus); Hs, human (Homo sapiens); Mm, mouse (Mus musculus); Lc, coelacanth (Latimeria chalumnae); Le, little skate (Leucoraja erinacea); Sc, catshark (Scyliorhinus canicula); Tr, pufferfish (Takifugu rubripes).