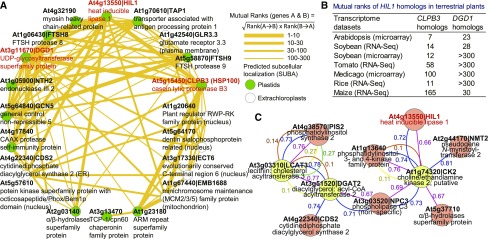
Figure 7.
The Gene Coexpression Network of HIL1 and the Genes Involved in Chloroplastic Abiotic Stress Responses.
(A) The gene coexpression network of HIL1 in Arabidopsis was calculated on the ATTED-II website. Genes predicted to be localized to chloroplasts are marked with green dots. The mutual ranks represent nonparametric values reflecting the order of correlated genes. The values ≤300 are shown.
(B) The coexpression of the HIL1, CLPB3, and DGD1 homologs was conserved among plant species. The values were obtained from the ATTED-II website.
(C) Coexpression analysis of the putative heat-inducible genes involved in glycerolipid metabolism. One hundred and eighty Arabidopsis microarray CEL files under heat stress were selected as a data set for the calculation (Supplemental Table 4). LCAT3, DGAT2, and CK2 (yellow circles) were searched as queries. Seven genes (red circles) were coexpressed with the queries, which were possibly induced by heat stress. The links represent a positive association and values, which were calculated from five different methods (Pearson, blue; Spearman, red; CLR, brown; MRNET, yellow; ARACNE, green). The result was visualized using Cytoscape software.