Figure 8.
Enzymatically Inactive pdNAD-MDH Proteins Can Complement the Embryo Lethality and Growth Defects of the pdnad-mdh Mutant.
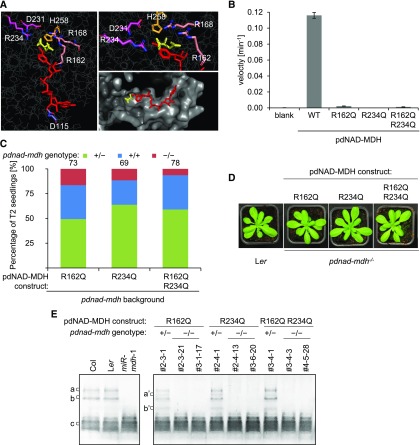
(A) Catalytic center of the Arabidopsis pdNAD-MDH structure. The pdNAD-MDH protein sequence (without cTP) was modeled using the human malate- and NADH-bound MMDH2 crystal structure (PDB: 2DFD) as a template. Left panel shows NADH (red) and malate (yellow), and the relative position of the conserved catalytic amino acid residues of pdNAD-MDH. A detailed view of the malate binding site is shown in the upper right panel. The surface of the catalytic pocket is shown in the lower right panel.
(B) An in vitro activity assay was performed using purified recombinant proteins. The proteins (10 µg) were incubated with an excess of cofactor (NADH) at 22°C. The reaction was started by the addition of excess substrate (oxaloacetate). The velocity was determined by measuring the decrease in absorbance at 340 nm resulting from the conversion of NADH to NAD+. Error bars indicate mean ± se (n = 3).
(C) Genotyping results of T2 progeny from T1 plants heterozygous for pdnad-mdh and expressing the enzymatically inactive pdNAD-MDH proteins. Numbers above the bars indicate the number of BASTA-resistant T2 plants that were genotyped.
(D) Photographs showing 3-week-old rosettes of homozygous pdnad-mdh plants complemented with enzymatically-inactive pdNAD-MDH mutants. For comparison, wild-type (Ler) plants are shown. Plants were grown under long days (16 h light/8 h dark).
(E) NAD-MDH activity observed by native-PAGE. Equal amounts of protein (15 µg) were loaded per lane, and all lanes were run on the same gel. pdNAD-MDH runs in distinct activity bands (a, b, and c). While the fastest migrating band (c) corresponding to the free dimer is masked by other NAD-MDH activity, the two slower migrating bands that correspond to NAD-MDH in protein complexes can be easily observed (a and b). Additional activity bands (a’ and b’) are observed for plants heterozygous for the pdnad-mdh T-DNA insertion expressing the catalytic-inactive pdNAD-MDH variants, possibly due to dimer formation between an inactive pdNAD-MDH with an endogenous form of pdNAD-MDH protein.