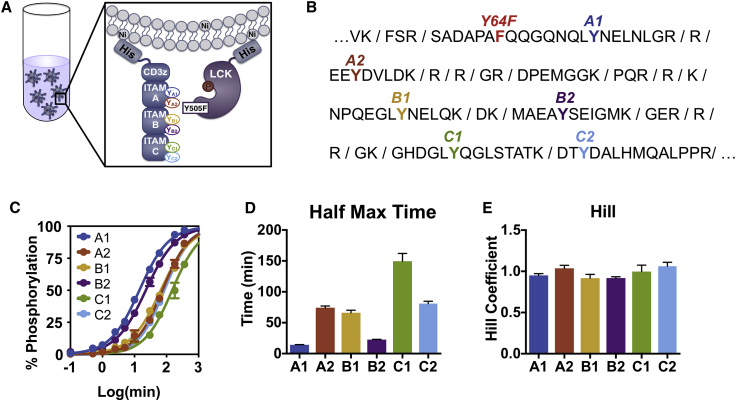
Figure 1.
CD3ζ sites are phosphorylated by LCK with different kinetics. (A) A schematic of the experimental liposomal system is shown. CD3ζ and LCK His-tagged proteins were purified and allowed to bind to large unilamellar liposomes bearing nickel-chelated lipids. Once proteins were bound, ATP was added, and the proteins were allowed to interact for various times before being subjected to phosphoproteomic mass spectrometry for quantification. (B) A sequence of CD3ζ intracellular domain is shown, with trypsin cut sites denoted. Individual ITAM tyrosine sites are labeled in different colors. Y64F indicates a tyrosine-to-phenylalanine mutation to ensure that each peptide only has one tyrosine phosphorylation site. This mutation does not influence overall phosphorylation kinetics (see Fig. S3). (C) Experimental data (circles) and sigmoidal fit (lines) for CD3ζ ITAM phosphorylation on liposomes containing 10% acidic POPS lipids are shown. Error bars represent the SD of two technical replicates normalized by site-specific standard curves. (D) The half-maximal time for each CD3ζ ITAM site is shown. Data represent mean and mean ± standard error of the fit to a four-parameter sigmoidal curve. (E) The Hill coefficient for each CD3ζ ITAM site is shown. Data represent mean and mean ± standard error of the fit to a four-parameter sigmoidal curve. To see this figure in color, go online.