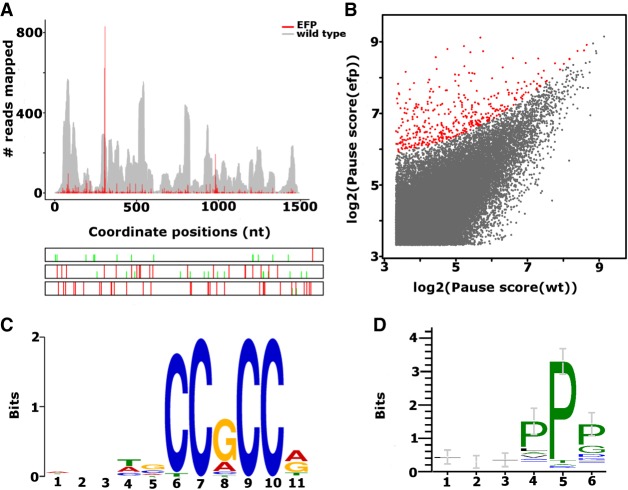
FIGURE 2.
(A) Rfeet visualization of the ribosome density profile (red) for the ubiD gene containing a ribosome pause detected by PausePred at position 307 of the CDS in an E. coli strain lacking elongation factor EFP (Woolstenhulme et al. 2015). The wild-type strain is provided as a gray coverage profile, i.e., where the number of sequence reads aligning to each coordinate is displayed. The bottom panel shows the ORF organization with red lines for stop codons and green lines for AUG codons. (B) Comparison of pause scores predicted by PausePred in E. coli mutants lacking elongation factor EFP and the wild-type sample. The EFP pauses with a pause score ≥50 compared to WT are highlighted in red. (C) The sequence motif found to be overrepresented in 88 cases, using 15 nt upstream of the detected pauses in the filtered 399 cases (B, highlighted in red). The pause score of these locations is higher than in the wild-type by at least 50. (D) The enrichment of the PP motif in the 88 sequences found to contain a C-rich motif at the nucleotide level.