Significance
How biological systems robustly sustain differentiated states is under active study. Here, we find that human cytomegalovirus, a herpesvirus that is a major cause of birth defects, encodes a transcriptional positive-feedback loop that temporally extends transient activation signals carried in the incoming viral particle to sustain the viral lytic expression cycle. Attenuation of this feedback loop severely impacts virus fitness, suggesting a new antiviral target that may extend to other herpesviruses.
Keywords: single-cell imaging, virus, feedback circuitry, mathematical model
Abstract
A fundamental signal-processing problem is how biological systems maintain phenotypic states (i.e., canalization) long after degradation of initial catalyst signals. For example, to efficiently replicate, herpesviruses (e.g., human cytomegalovirus, HCMV) rapidly counteract cell-mediated silencing using transactivators packaged in the tegument of the infecting virion particle. However, the activity of these tegument transactivators is inherently transient—they undergo immediate proteolysis but delayed synthesis—and how transient activation sustains lytic viral gene expression despite cell-mediated silencing is unclear. By constructing a two-color, conditional-feedback HCMV mutant, we find that positive feedback in HCMV’s immediate-early 1 (IE1) protein is of sufficient strength to sustain HCMV lytic expression. Single-cell time-lapse imaging and mathematical modeling show that IE1 positive feedback converts transient transactivation signals from tegument pp71 proteins into sustained lytic expression, which is obligate for efficient viral replication, whereas attenuating feedback decreases fitness by promoting a reversible silenced state. Together, these results identify a regulatory mechanism enabling herpesviruses to sustain expression despite transient activation signals—akin to early electronic transistors—and expose a potential target for therapeutic intervention.
A common problem in information-transmission systems is how to robustly maintain signal strength despite the inevitable signal decay that occurs through any transmission medium. For example, in the last century, the need to reamplify telephone signals, which diminished in strength over transmission-line distance, spurred innovations, such as vacuum-tube repeaters and point-of-contact and junction transistors (1, 2). In developmental biology, a corresponding robustness problem regards how developmental states are stably maintained, often referred to as “canalization” for how the “canal” walls in an epigenetic landscape stabilize the developmental state (3). During canalization, biological systems must overcome the signal-decay problem and stably maintain a developmental state long after degradation of initiating signals and various mechanisms evolved to convert transient signals to sustained outputs (4–9). The fundamental issue facing biological signaling molecules, often proteins or nucleic acids, is that they have intrinsic degradation times mediated either by environmental mechanisms (e.g., UV radiation) or active intracellular machinery (e.g., the proteasome). The mechanisms allowing cells to sustain differentiated phenotypes long after degradation of the initial (transient) differentiation signals remain actively studied (5, 7).
Viruses face a version of the canalization problem as they balance a temporal trade-off: On the one hand, a virus must rapidly counteract cellular defenses to initiate its lytic cycle, but on the other hand the virus must sustain repression of cellular defenses over the course of its intracellular viral life cycle, which can last for many hours or even days. To antagonize cellular defenses as rapidly as possible after infection, herpesviruses have evolved to carry transactivator molecules within the infecting virion particle and thereby circumvent the delays inherent to de novo gene expression. However, these transactivator molecules are immediately subject to cellular degradation mechanisms upon infection, and herpesvirus life cycles can extend for days (10). How, or if, these transactivator signals are sustained across the course of the viral life cycle remains unknown. Beta herpesviruses, having intracellular life cycles lasting over 4 d, appear to have a particularly large transient-versus-sustained signaling problem to overcome.
The beta herpesvirus human cytomegalovirus (HCMV)—a leading cause of birth defects and transplant failures—must rapidly overcome innate host-cell defense mechanisms to initiate its lytic cycle. Cellular promyelocytic leukemia nuclear bodies (PML-NBs; a.k.a. nuclear domain 10, ND10) (11)—containing PML, the death domain-associated protein (DAXX), and SP100—act to repress viral gene expression and prevent HCMV from initiating its lytic cycle (12–14). To counteract these cellular defenses, HCMV virions package the viral transactivator phosphoprotein 71 (pp71) in the virion’s tegument layer (15). pp71 is the major tegument transactivator protein and interacts with DAXX to promote its degradation. Upon infection, pp71 tegument proteins are released into the cell, disrupt PML-NBs to antagonize the cellular silencing machinery, and activate the HCMV major immediate-early promoter (MIEP). The MIEP initiates the viral transcription program, and its main protein products, the immediate-early 1 (IE1) and 2 (IE2) proteins, are obligate for lytic expression by regulating host and viral genes (16–19), manipulating the host cell cycle (20, 21) and circumventing host innate defense (22, 23). Whereas IE2 is obligate at all multiplicities of infection (MOIs), IE1 appears to be obligate only at low MOI (11, 24–26). We asked how HCMV is able to sustain the pp71 transactivation signal—which is inherently transient due to pp71 proteolysis—to avoid cell-mediated silencing over an extended 96-h cellular infection cycle.
Building off theoretical analysis showing that autoregulatory circuitry (i.e., feedback) can promote cellular “memory” to transient differentiation signals during developmental canalization (3, 5), we hypothesized that positive feedback may enable the virus to extend a transient transactivation signal. Positive feedback, whereby a signaling molecule autoactivates its own expression or drives a downstream autoactivating loop, can occur through a variety of mechanisms, including transcriptional transactivation, enhanced translation, and even reduced degradation (for a review see ref. 5). Such feedback motifs are widely used in biological systems to execute cell-fate decisions in both unicellular and multicellular organisms (5, 8, 27–32). HCMV encodes a strong immediate-early negative-feedback loop to maintain IE2 levels below a cytotoxic threshold (33), but whether HCMV encodes an immediate-early positive feedback loop that could extend transient tegument transactivation signals is unclear. One possibility is that IE1, which can weakly transactivate the MIEP (34), could encode positive-feedback circuitry to temporally extend the duration of MIEP transactivation, but whether this autoregulation was strong enough to be physiologically relevant was not clear.
Here, through the use of mathematical modeling, we develop an assay to probe IE1 positive-feedback strength and its physiological relevance to viral replication. To test the feedback-mediated temporal extension hypothesis, we constructed a tunable, two-color HCMV reporter virus where both IE1 and IE2 can be followed in real time and IE1 proteolysis can be pharmacologically tuned using a small molecule via a degron tag (35). Using single-cell time-lapse imaging (Movies S1 and S2), we find that IE1 positive feedback is sufficiently strong to convert transient pp71 transactivation into sustained lytic expression. The results show that IE1 positive feedback is essential for sustained MIEP expression and that attenuating the viral positive-feedback strength causes a severe fitness defect by promoting entry to a silenced state. These findings suggest that transcriptional positive feedback in herpesviruses acts as a transistor-like mechanism to temporally extend intrinsically transient tegument signals and facilitates canalization of the productive (lytic) viral expression state.
Results
The HCMV Major Tegument Transactivator Has an 8-h Lifetime but Modeling Predicts That a Putative Positive-Feedback Loop Could Sustain Transactivation.
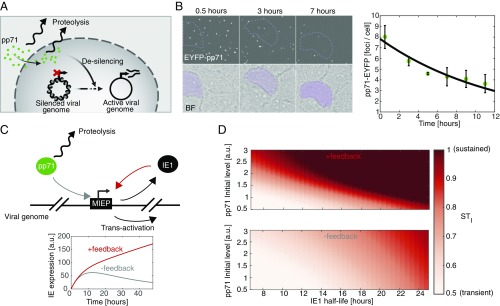
To understand how HCMV can sustain expression from the MIEP despite stable cellular silencing (Fig. 1A), we first measured the degradation rate of the major tegument transactivator pp71 (24) after infection. ARPE-19 cells were synchronously infected with a recombinant HCMV encoding an EYFP-pp71 fusion (36) and followed for 11 h using time-lapse fluorescence microscopy. pp71 appeared as distinct fluorescent foci within cells with the number of foci decreasing during the first few hours of infection (Fig. 1B). Quantifying the temporal kinetics of the number of pp71 foci per cell results in a measured pp71 half-life of 8.1 ± 1.1 h (Fig. 1B), which agrees with cycloheximide-based estimates of the decay rate of total cellular pp71 (SI Appendix, Fig. S1 A and B). Moreover, we were unable to detect cleavage products of the enhanced YFP (EYFP)-pp71 fusion protein in infected cells, indicating that EYFP signal decay likely reflects pp71 decay (SI Appendix, Fig. S1C). Nevertheless, the 8-h pp71 half-life we measured was used as an upper limit, since it is possible that fusion of EYFP to pp71 increases its stability (36).
Fig. 1.
Tegument-derived pp71 is transient in cells, but modeling predicts that positive feedback could sustain transactivation. (A) Schematic of initial regulatory events occurring during infection of cells with HCMV. (B) Time-lapse fluorescence microscopy of ARPE-19 cells synchronously infected with an EYFP-pp71 HCMV virus (AD169) at MOI of 1 and quantification of pp71 cellular levels over the first 11 h of infection (an average of 162 cells). (C and D) A mathematical model describing pp71 and IE1 dynamics. Eqs. 1 and 2 were solved numerically, using ODE45 solver (MATLAB; MathWorks). STI was calculated for every set of parameters as the ratio between IE1 expression at 24 h after infection and maximal IE1 expression. For parameters, see SI Appendix, Table S2.
We next used mathematical modeling to simulate the expected MIEP activity given this measured pp71 decay rate. Since IE1 is transcribed from the MIEP during the first few hours of infection, and, similar to pp71, IE1 localizes to PML bodies to antagonize these cellular antiviral defenses (22, 37, 38), we hypothesized that IE1 may play a role in sustaining MIEP expression. To examine the theoretical validity of this hypothesis (Fig. 1C), we constructed a highly simplified ordinary differential equation (ODE) model of IE1 expression in the presence of a decaying pp71 transactivation signal using a “course-grained” modeling approach to generate the following nonlinear ODE model:
| [1] |
| [2] |
Importantly, the course-grained modeling approach is not intended as a comprehensive description of all known molecular interactions. Instead, the goal is to find a minimum set of interactions and components capable of generating testable predictions about the duration of IE expression—for this reason, the IE2 negative-feedback accelerator circuitry (33) was determined to be expendable (SI Appendix, Fig. S2 and Table S3). In this highly simplified, course-grained model, only “active” pp71 and IE1 protein levels are considered, and we attempt to limit the number of unmeasured parameters. The pp71 decay, per-capita rate γ1, is measured above, and the per-capita IE1 proteolysis rate γ2 was previously measured (39). The model also accounts for IE transactivation by pp71 and IE1 (Eq. 2); α0 represents the basal rate of MIEP-driven IE1 expression, α1 represents the transactivation of the MIEP by pp71, and α2 accounts for IE1 transactivation of the MIEP and is the putative positive-feedback gain parameter. k1 and k2 are Michaelis-like constants for transactivation, and h1 and h2 are Hill coefficients to account for self-cooperativity of transactivation. Simulations of a more detailed model, which includes the IE2 negative-feedback accelerator circuit (33), yield similar results and qualitatively match the temporal dynamics of IE2 expression observed in single cells (SI Appendix, Fig. S2).
As expected, simulations of Eqs. 1 and 2 show that the temporal duration of IE expression is significantly affected by the IE1 degradation rate (Fig. 1C). When the IE1 half-life is long (∼24 h in the wild-type case as measured below), IE expression is sustained and monotonically increasing over the first 24 h. However, when IE1 half-life is reduced, IE expression becomes transient with a dramatic drop during the first 24 h postinfection. When positive feedback is removed by abrogating IE1 transactivation of the MIEP (α2 set to zero), IE expression does not exhibit sustained expression for either long or short IE1 half-life (Fig. 1C).
To quantify the duration of IE expression, we also defined a sustained-versus-transient IE-expression index, STI—the ratio between IE1 expression level at 24 h postinfection and the maximal IE1 expression peak in an individual cell (essentially a version of the second derivative test, used here as a canalization measure). When a cell’s STI is high (>0.5), the cell is able to sustain at least 50% of peak IE-expression levels, whereas when a cell’s STI is low (<0.5), the cell is unable to sustain IE expression and it drops below 50% of its peak value. We examined STI over a wide range of parameters and, as expected, the STI was significantly affected by both the IE1 degradation rate and the initial level of pp71 at the time of infection (Fig. 1D). These simulations showed that when IE1 degradation is slow, IE expression is sustained (high STI) even when initial pp71 levels are low (Fig. 1D, Top). However, in the absence of IE1 positive feedback (α2 set to zero), IE expression is transient, and a much longer IE1 half-life (i.e., super physiological) is required to sustain IE expression (Fig. 1D, Bottom).
These results suggested that IE1 positive feedback acts to extend the duration of pp71 transactivation and proposed an assay for the presence of IE1 positive feedback, based on movement along the horizontal axis in Fig. 1D. If IE1 positive feedback is sufficiently strong, reducing the IE1 half-life (right to left in Fig. 1D) should switch IE expression profiles (both IE1 and IE2) from sustained to transient (high to low STI). In contrast, if positive feedback is weak or absent, reducing the IE1 half-life (within the physiological range) will have little qualitative effect on the sustained-versus-transient expression profiles. In addition, the simulations predicted that high initial levels of pp71 could also sustain IE expression, even in the absence of feedback (Fig. 1D).
Tuning IE1 Degradation Reveals a Positive-Feedback Loop Required to Sustain HCMV Immediate-Early Gene Expression.
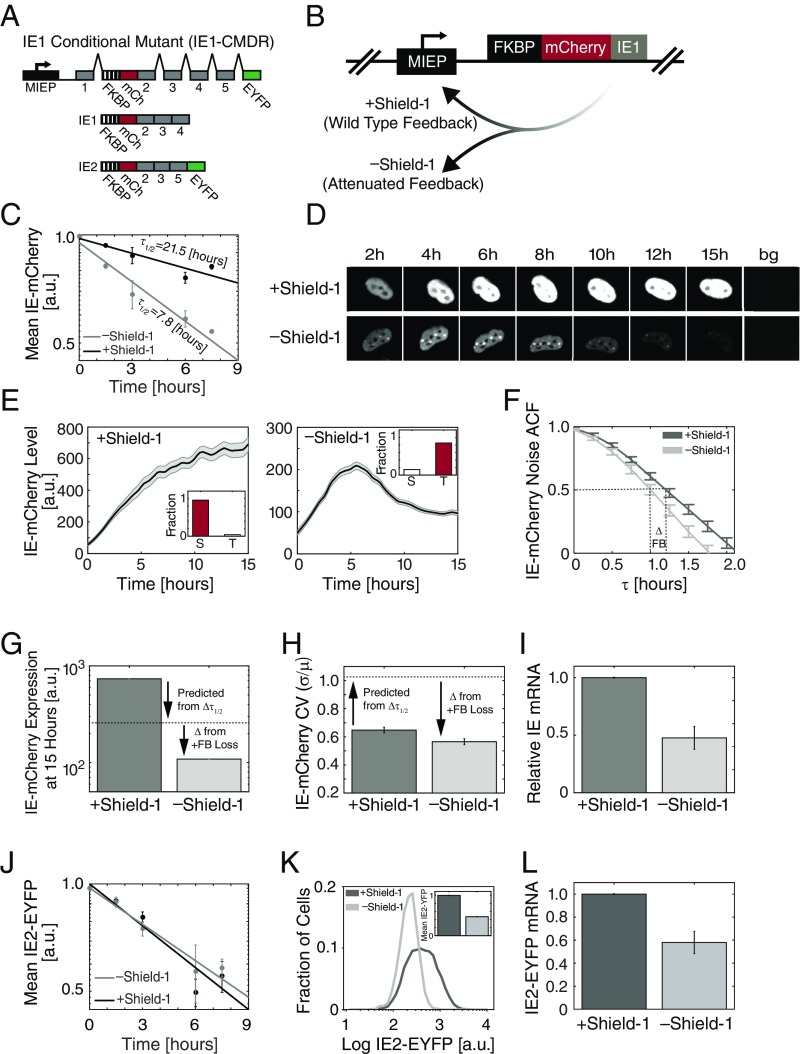
To test the model predictions on the role of IE1 positive feedback in sustaining IE expression, we generated an IE1-conditional mutant dual-reporter virus (HCMV-IE1-CMDR). First, on the background of a previously characterized TB40e-IE2-EYFP parent virus (33), we fused an mCherry reporter to the N terminus of IE1 (automatically also tagging IE2) to enable real-time tracking of the immediate-early promoter dynamics during infection (Fig. 2 A and B); we could not detect cleavage products of the mCherry-IE fusion in infected cells (SI Appendix, Fig. S3A), indicating that mCherry signal likely reflects IE expression levels. Second, to tune the IE1 degradation rate, we fused an FKBP degron tag (35) to the N terminus of mCherry-IE1. In the presence of the small molecule Shield-1, the IE1 degradation rate is unperturbed by the FKBP degron, but in the absence of Shield-1 the tagged protein is rapidly degraded. This N terminus tagging approach for IE1 was required since attempts to fuse proteins to the C terminus of IE1 generated virus with MOI-dependent replication defects. In addition to the N terminus IE1 tags, this reporter virus has EYFP fused to the C terminus of IE2, so that the IE2 protein can be tracked in parallel to IE1. Since IE1 is far more abundant than IE2 (39) and has a substantially longer half-life, mCherry primarily reports on IE1, with minimal contribution from IE2. In the presence of Shield-1, this IE1-CMDR virus grows with replication kinetics similar to those of the parent virus (SI Appendix, Fig. S3B).
Fig. 2.
IE1 positive feedback sustains expression from the HCMV immediate-early promoter. (A) Map of the MIE region of the recombinant HCMV TB40/E IE1 conditional-mutant dual-reporter virus (IE1-CMDR) showing the MIEP (black) and IE region exons 1–5 (gray), along with the recombinant fusions: Exon 2 is genetically fused at its N terminus to mCherry (mCh, red) and the FKBP degron tag (hashed) that destabilizes the IE1 protein in the absence of the small molecule Shield-1; exon 5 also contains a fusion to EYFP (green) at its C terminus. Translation typically begins at the 5′ end of exon 2 (now FKBP). See also SI Appendix, Fig. S3. (B) Schematic of the putative effects of Shield-1 on IE1-mediated positive feedback. (C) Degradation rate of IE1 in cells infected with IE1-CMDR virus as measured by flow cytometry. Protein synthesis was blocked with cycloheximide at 24 h after infection (time = 0) and the degradation rate was measured ±1 μM Shield-1. Shown is the average of two repeats, and error bars denote SD. Decay rate was calculated by fitting the data to an exponential decay model (solid lines). (D) Representative images from fluorescence time-lapse microscopy of IE-mCherry expression in cells infected with IE1-CMDR virus. Cells were cultured in medium containing 1 μM Shield-1 (Top) or without Shield-1 (Bottom). Cells were cultured in medium with 1 μM Shield-1 (Top, wild-type feedback) or without Shield-1 (Bottom, attenuated feedback) and tracked over time; “bg” represents background autofluorescence image. (E) IE-mCherry expression of cells infected with IE1-CMDR virus. Cells were cultured in medium with 1 μM Shield-1 (Left, wild-type feedback, averaged over 119 cells) or without Shield-1 (Right , attenuated feedback, averaged over 153 cells). Bold line denotes mean (i.e., general trend) of the population with gray shading showing SE. Cell trajectories were digitally synchronized to the first detection of mCherry signal. (Inset) The fraction of cells with sustained (S, STI > 0.5) or transient (T, STI < 0.5) IE-mCherry expression over three biological repeats. (F) HF-ACF of cells infected with IE1-CMDR virus in the presence of 1 μM Shield-1 (dark gray) or without Shield-1 (light gray). Shown is an average over 100 cells each; error bars denote SE. (G and H) IE-mCherry mean expression level and noise (CV) 15 h after infection. Dashed line is the expected value from change in IE1 half-life alone (Δ from τ1/2), whereas the additional difference (Δ) is ascribed to the loss of IE1 positive feedback. (I) Relative IE-mCherry mRNA levels (from RT-qPCR) in cells infected with IE1-CMDR virus ±1 μM Shield-1 (P < 0.01, two-tailed t test). (J) Degradation rate of IE2-EYFP in cells infected with IE1-CMDR virus as measured by flow cytometry. Protein synthesis was blocked with cycloheximide at 24 h after infection (time = 0) and degradation rate was measured ±1 μM Shield-1. Shown is the average of two repeats, and error bars denote SD. Decay rate was calculated by fitting the data to an exponential decay model (solid lines). (K) Flow cytometry for IE2-EYFP levels in cells infected with IE1-CMDR virus. Cells were cultured in medium with 1 μM Shield-1 or without Shield-1. (Inset) Normalized mean fluorescence of IE2-EYFP. (L) Relative IE2-EYFP mRNA levels (from RT-qPCR) in cells infected with IE1-CMDR virus ±1 μM Shield-1 (P = 0.012, two-tailed t test).
To measure the degradation rate of IE1 protein, the decay in mCherry intensity was quantified after cycloheximide translation block in the presence and absence of 1 μM Shield-1 24 h after IE1-CMDR infection of ARPE-19 cells. The results show that the FKBP degron decreases IE1 protein half-life from 21.5 to 7.8 h, or approximately threefold (Fig. 2C and SI Appendix, Fig. S4).
Next, we used single-cell resolution time-lapse microscopy to track IE-expression dynamics in ARPE-19 cells infected with the HCMV IE1-CMDR virus. In cells infected in the presence of 1 μM Shield-1 (+Shield-1), IE gene expression from the MIEP reached steady state ∼10 h after infection and remained at steady state (STI > 0.5) in >95% of cells (Fig. 2 D and E and Movie S1). These dynamics are in temporal agreement with previous results using the HCMV AD169 laboratory strain (33). Strikingly, infection in the absence of Shield-1 (−Shield-1) results in transient IE expression (STI < 0.5) of IE1 (Fig. 2 D and E and Movie S2) in most cells (>80%). This steep drop in IE1 levels in the absence of Shield-1 occurs despite IE expression initiating at a similar time compared with the +Shield-1 condition.
To quantify changes in feedback strength, we used high-frequency autocorrelation function (HF-ACF) analysis. ACF analysis is a common signal-processing technique for time-lapse data that analyzes the frequency of fluctuations, with changes in half-correlation time (ct50) of fluctuation used to determine biophysical properties (e.g., in fluorescence spectroscopy) (40, 41). Shifts in the ct50 of the HF-ACF are a sensitive reporter of feedback strength, and the HF-ACF is largely unaffected by changes in protein half-life (40, 41). When HF-ACF of the IE1-mCherry single-cell trajectories is analyzed (Fig. 2F), it shows that IE1 ct50 is significantly reduced when Shield-1 is removed, indicating attenuation of positive feedback (40, 41).
This IE1 positive feedback, in fact, appears to be required to account for the observed changes in IE mean-expression level and variance. Specifically, the observed change in IE1 level when Shield-1 is removed is substantially greater than can be accounted for by a threefold change in IE1 half-life (Fig. 2G). In the absence of positive feedback, a threefold decrease in IE1-mcherry half-life should confer a threefold decrease in IE1-mcherry steady-state levels. However, IE-mCherry levels 15 h after infection show a sevenfold decrease. This more than threefold change in expression level is consistent with attenuation of an IE1 positive-feedback loop that was acting to amplify MIEP expression. Likewise, the change in the magnitude of IE expression fluctuations measured by the coefficient of variation (CV) of the trajectories cannot be accounted for by the change in IE1 half-life alone, based on Poisson scaling (Fig. 2H); specifically, for a simple birth–death Poisson process, a sevenfold decrease in mean-expression level should correspond to an ∼2.65-fold increase in CV (42). However, the data show that this sevenfold decrease in mean IE1 expression levels generates a decrease in the IE1 CV (Fig. 2G). This decrease in IE1 CV despite decreased mean is consistent with attenuation of positive feedback (6).
To directly test if IE1 destabilization influences MIEP expression, we quantified IE mRNA (as opposed to protein) in the presence and absence of Shield-1 by RT-qPCR (Fig. 2I). We observed an approximately twofold decrease in MIEP activity upon Shield-1 removal (Fig. 2I), indicating that reduction in IE1’s half-life confers a change in IE mRNA levels, and hence an attenuation of the positive feedback strength. This twofold change in IE mRNA levels accounts for the missing quantitative difference (Δ) in the IE protein levels unaccounted for by the IE1 half-life change (Fig. 2G).
We also analyzed the effect of IE1 destabilization on IE2 levels. Notably, despite the FKBP tag also being present on the IE2 protein, the degradation rate of IE2 protein does not appear affected by Shield-1 removal (Fig. 2J), presumably due to IE2’s relatively short native half-life (33). Despite IE2-EYFP’s half-life’s not being affected by the FKBP degron tag, IE2-EYFP expression exhibits a twofold decrease in abundance upon infection in the absence of Shield-1 compared with infection in presence of Shield-1 (Fig. 2K and SI Appendix, Fig. S5D). This twofold change in IE2 protein levels corresponds to an approximately twofold decrease in IE2-EYFP mRNA levels (Fig. 2L). We also analyzed the HF-ACF for IE2-EYFP—whose half-life remains unchanged by Shield-1—and found a ct50 shift similar to the shift in IE1 ct50 (SI Appendix, Fig. S5), further validating the presence of an IE1 positive-feedback loop.
Overall, these single-cell imaging data, of both IE1 and IE2, indicate the presence of an IE1 positive-feedback loop that is attenuated when IE1 half-life is reduced by Shield-1 removal.
Attenuating IE1 Positive Feedback Decreases Viral Fitness by Promoting Entry to a Reversible Silenced State.
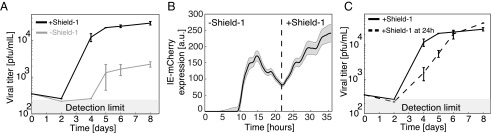
Next, to determine if sustained IE expression is required for efficient virus replication and, correspondingly, if transient IE expression results in a replicative fitness defect, we measured HCMV viral replication in the setting of both wild-type IE1 feedback (+Shield-1) and attenuated IE1 feedback (−Shield-1). ARPE-19 cells were infected with the IE1-CMDR virus (MOI = 0.02) in the presence of Shield-1 (wild-type feedback) and absence of Shield-1 (attenuated feedback), and resulting virus titers were assayed over time by TCID-50. The viral replication kinetics revealed an ∼10-fold reduction in viral titer when IE1 positive feedback was attenuated (−Shield-1), compared with the wild-type IE1 feedback case (+Shield-1) (Fig. 3A). Although these data do not suggest the presence of positive feedback (which is shown in Fig. 2), they indicate that sustained IE expression confers replicative fitness for HCMV.
Fig. 3.
Attenuated IE1 positive feedback decreases replication fitness, but feedback can be rescued to restore fitness. (A) Viral titers from cells infected with IE1-CMDR virus. Cells were cultured in media with 1 μM Shield-1 (black, wild-type feedback) or without Shield-1 (gray, attenuated feedback). Virus was harvested at different time points after infection. Viral titers were measured in medium containing 1 μM Shield-1; error bars denote SD. (B) IE-mCherry expression from time-lapse single-cell microscopy of cells infected with IE1-CMDR virus. Cells were cultured in medium without Shield-1 for 24 h and then supplemented with 1 μΜ Shield-1 for 12 h. Bold line denotes mean mCherry expression (67 cells); gray shading denotes SE. (C) Viral titers from cells infected with IE1-CMDR virus. Cells were cultured in medium with 1 μM Shield-1 (solid line), or Shield-1 was added to the medium after 24 h (dashed line). Virus was harvested at different time points after infection, titers were measured in media containing 1 μM Shield-1, and error bar denotes SD.
We next asked if the IE-expression transient led to abortive infection or instead promoted a latent-like, silenced state that could be reactivated if IE expression was rescued at a later time. If Shield-1 was added back 24 h after infection, IE expression kinetics could be restored to a sustained trajectory (Fig. 3B), and correspondingly viral replication could be rescued to wild-type levels (Fig. 3C).
High pp71 Abundance Partially Rescues Attenuation of the IE1 Positive-Feedback Loop.
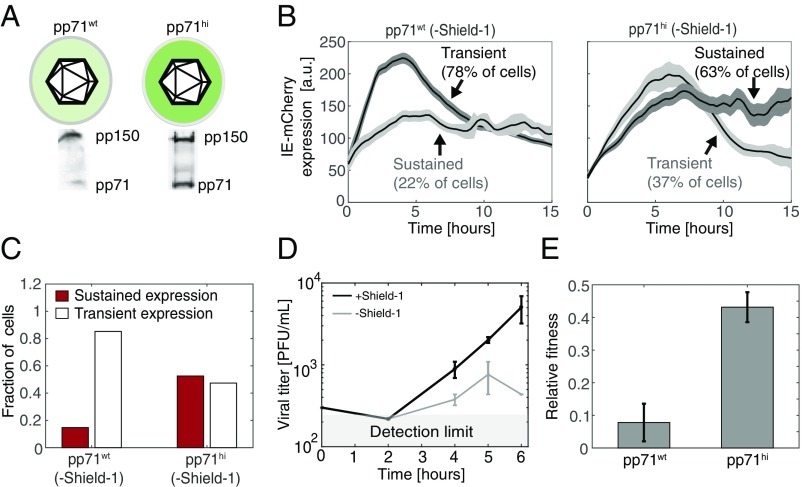
As a further test of the model, we examined the second prediction of Eqs. 1 and 2 that high initial abundance of pp71 could compensate for weak IE1 positive feedback to enable sustained IE expression. To test this prediction, we artificially increased the pp71 tegument load in HCMV virion particles by packaging the IE1-CMDR virus on a pp71-expressing cell line (24). We reasoned that ectopic expression of pp71 from the cell line in addition to the pp71 expressed from the viral genome would result in packaging of more pp71 into virions. Comparing pp71 levels between the two purified viral preparations (pp71hi and pp71wt) by Western blot showed about a threefold increase in pp71 abundance in the pp71hi virus isolate (Fig. 4A).
Fig. 4.
Increased pp71 abundance partially rescues expression and fitness of attenuated feedback virus. (A) Western blot analysis of pp71 in viral particles packaged on cells overexpressing pp71 (IE1-CMDR-pp71hi) or on nonoverexpressing cells (IE1-CMDR-pp71wt). (B) Representative time-lapse microscopy traces of IE-mCherry expression in cells infected with IE1-CMDR-pp71hi or IE1-CMDR-pp71wt. Cells were cultured without Shield-1 (attenuated feedback) and classified as sustained or transient based on expression kinetics as defined above. Bold line denotes mean mCherry signal of sustained or transient cells; shaded area denotes SE. Cell trajectories were digitally synchronized to the first detection of mCherry signal. (C) The fraction of cells with sustained IE-mCherry expression (STI > 0.5) after infection with IE1-CMDR-pp71hi virus (116 cells) or IE1-CMDR-pp71wt virus (243 cells). Cells were cultured without Shield-1 (attenuated feedback). P < 0.001 was calculated using a Fisher exact test. (D) High pp71 can partially compensate the fitness lost when feedback is attenuated. Viral titers from cells infected with IE1-CMDR-pp71hi virus ± Sheild-1. Cells were infected in medium with 1 μM Shield-1 (black, wild-type feedback) or without Shield-1 (gray, attenuated feedback), virus was harvested at indicated times after infection, and titers were measured in medium supplemented with 1 μM Shield-1. Compare with Fig. 3A. (E) High pp71 partially compensates for fitness lost when feedback is attenuated. Relative single-round fitness (titer at day 4) for either IE1-CMDR-pp71wt (Left) and IE1-CMDR-pp71hi (Right) when feedback is attenuated (−Shield-1) normalized to viral titer for corresponding wild-type feedback (+Shield-1) case (P = 0.041, two-tailed t test).
Next, we followed IE expression in cells infected with either IE1-CMDR-pp71hi or IE1-CMDR-pp71wt in an attenuated feedback setting (i.e., −Shield-1) using time-lapse microscopy. Strikingly, when cells are infected with the IE1-CMDR-pp71hi virus, the percentage of infected cells exhibiting sustained IE gene expression is >50% (Fig. 4 B and C), despite the absence of Shield-1 (i.e., attenuated feedback), whereas only ∼20% show sustained IE expression when infected with the IE1-CMDR-pp71wt virus in the absence of Shield-1. Thus, the results appear to validate the model prediction that increased pp71 can compensate for attenuated feedback to sustain IE expression kinetics.
Next, we used these results to estimate the relative contribution of IE1 and pp71 to the activation of the MIEP. To this end, we created a stochastic version of the course-grain model (SI Appendix, Fig. S6 and Table S4). We varied IE1 and pp71 transactivation levels and examined which regime of parameters yields an increase in the percentage of stable cells from 20 to 50% when the initial pp71 levels increase by threefold. The results suggest that to satisfy these constraints MIEP activation by pp71 should be similar to its activation by IE1 (SI Appendix, Fig. S6); if autoactivation by IE1 were substantially stronger than pp71 per-capita activation, the simulations predict that the probabilities of stable cells would be much greater than experimentally observed.
Finally, we checked if pp71’s partial rescue of sustained IE expression in the attenuated-feedback setting also translated into a rescue of viral fitness. Cells were infected at low MOI (0.02) with the IE1-CMDR-pp71hi virus in the presence or absence of 1 μM Shield-1 (i.e., wild-type feedback and attenuated feedback, respectively), and resulting virus titers were assayed over time by TCID-50 (Fig. 4D). For the pp71hi infection, viral titers decrease twofold 4 d postinfection, when feedback is attenuated (Fig. 4E). Compared with the >10-fold decrease in fitness when feedback is attenuated in pp71wt infection, this twofold decrease in fitness in the pp71hi setting is substantially less (Fig. 4E), indicating that boosting initial pp71 levels can partially compensate for attenuated IE1 positive feedback to mitigate the viral fitness defect.
Overall, these results suggest that the duration of IE expression is important for viral replication and that IE1 positive feedback converts inherently transient pp71 transactivation signals into sustained IE expression profiles to drive efficient viral replication.
Discussion
Herpesviruses face a temporal trade-off between the need to rapidly counteract cellular defenses and initiate lytic infection and the need to sustain the resulting lytic expression patterns during infection. This balancing appears to have resulted in a transient-signaling dilemma as the most rapid mechanism to initiate transactivation is to circumvent de novo gene expression and package mature transactivator molecules directly within the infecting virion particle, but doing so subjects transactivator molecules to degradation (without de novo production) immediately upon infection of the cell. Beta herpesviruses, such as HCMV, must overcome a particularly severe transient-signaling problem since their infection cycle lasts for days, but the half-life of its major transactivation signal (pp71) is hours (Fig. 1). Mathematical modeling predicted that a putative positive-feedback circuit encoded by HCMV’s IE1 could sustain HCMV MIEP expression despite pp71 degradation (Fig. 1) and suggested an assay to probe for IE1 positive feedback. This assay relied on altering the IE1 degradation rate, which required construction of a conditional-mutant dual-reporter IE1 virus (IE1-CMDR). Time-lapse imaging of IE1-CMDR indicated the presence of IE1 positive-feedback circuitry (Fig. 2 and Movies S1 and S2) and showed that this IE1 positive-feedback circuit confers replicative fitness on the virus (Fig. 3).
One technical caveat is that the pp71 half-life we measured (8.1 ± 1.1 h) is based on the decay of the EYFP-pp71 fluorescence signal after infection of cells with EYFP-pp71 virus. Since it is possible that fusion of EYFP to pp71 increases pp71 half-life (36), 8 h serves as an upper limit for pp71 half-life. However, if the pp71 half-life is in fact shorter than 8 h, the need for a mechanism (e.g., positive feedback) that compensates for pp71 decay is even more pronounced.
Interestingly, the mechanisms of action by which pp71 and IE1 antagonize silencing from the PML bodies are different: pp71 induces SUMOylation and degradation of DAXX (26), while IE1 blocks PML SUMOylation, leading to dispersion of the PML bodies (43). Nonetheless, the data herein reveal a functional overlap between these different mechanisms of action: While IE1 positive feedback sustains expression from the MIEP during pp71 degradation, boosting pp71 levels in the virion can partially rescue MIEP expression and viral replicative fitness despite attenuated IE1 positive feedback (Fig. 4). These data suggest that IE1 activity should reach a certain threshold at early times of infection—to facilitate efficient viral replication—and that IE1 might be dispensable at late times of infection, as it can be partially complemented by high levels of pp71 at early times.
A limitation of the model we developed (Eqs. 1 and 2) is that it focuses exclusively on the early times after infection. At later times, different IE2 isoforms such as IE2 40 and IE2 60 are expressed. While these isoforms are not essential for viral replication they appear to have a contribution to viral fitness as their deletion affects the expression of other viral genes such as pp65 and the DNA replication factor UL84 (19, 44). Hence, it is possible that some of the difference in viral fitness we detect in the absence of Shield-1 (Fig. 3A) is due to a decrease in the abundance of these other IE2 isoforms.
Intriguingly, this feedback circuitry bears some resemblance to the latency control circuit in HIV-1, where a decision between proviral latency and active replication is regulated by the Tat transcriptional positive-feedback loop (45). However, there are important mechanistic differences as there is no reported tegument equivalent in HIV-1. Instead, the viral integration site of the HIV-1 provirus determines the basal activity of the viral promoter, and Tat directly autoactivates its own promoter (the LTR) without acting through PML disruption. It is also not clear if HIV-1 faces a transient-versus-sustained signaling trade-off as in the herpesviruses since the chromatin-based silencing mechanisms in T cells occur on the order of days to weeks, whereas the HIV-1 life cycle is completed within about 40 h (46, 47). Nevertheless, the fact that a positive-feedback loop controls the decision between active and latent replication in such an evolutionarily distant virus points to the potential generality of such feedback loops in regulating viral latency establishment.
From an evolutionary standpoint, it is interesting to speculate as to why pp71 did not evolve to directly autoregulate its own expression (i.e., without the need for the additional IE1 circuit “node”). A possible answer is that pp71 expression appears to be cytotoxic, as ectopic pp71 expression requires a late promoter (24, 48). So, one possibility is that early pp71 de novo expression could result in cytoxicity for the cell and, hence, IE1 evolved as a complementary mechanism to sustain the initiation of lytic gene expression and circumvent such cytotoxicity.
Overall, this study illustrates how positive-feedback circuitry is essential to sustain a viral lytic expression program and the associated fitness of a herpesvirus. These findings suggest a potentially broad mechanism for sustaining gene expression triggered by a transient signal that might extend to other herpesviruses, such as HSV-1 and EBV, that use tegument proteins to overcome host immune defenses and initiate the viral lytic cycle (24, 49, 50). As such, this mechanism may provide a new set of drug targets for development of herpesviruses antivirals.
Materials and Methods
Cell Culture, Viruses, and Media.
ARPE-19 cells (ATCC CRL-2302) were grown in DMEM/F-12 50/50 (Corning 10-090-CV) medium supplemented with 10% FBS and 50 U/mL penicillin–streptomycin at 37 °C and 5% CO2 in a humidified incubator.
WF28 cells (24) and life-extended human foreskin fibroblasts (HFFs) were grown in DMEM (Corning 10-013-CV) medium supplemented with 10% FBS and 50 U/mL penicillin–streptomycin at 37 °C and 5% CO2 in a humidified incubator.
Tb40/E-IE1 CMDR virus was expanded on ARPE-19 or WF28 cells in the presence of 1 μM Shield-1 for 3 wk until 100% cytopathic effect (CPE) was detected. Virus was harvested and then concentrated on a 20% sorbitol cushion (WS28 ultracentrifuge, 71,934 × g for 2 h). The pellet was resuspended in 1 mL of fresh medium and titered on ARPE-19 cells by TCID-50 to determine concentration of the viral stock.
EYFP-pp71 virus was expanded on HFFs for 3 wk or until 100% CPE was detected, harvested as described above, and titered on HFFs by TCID-50 to determine concentration of the viral stock.
Time-Lapse Microscopy.
ARPE19 cells (5 × 104 per well) were seeded in an eight-well imaging chamber (155409; Lab-Tek) and grown to confluency for 3 d. HCMV infections were synchronized on ice (MOI = 1), virus was removed after 30 min, and the plate was placed under the microscope for subsequent imaging. All imaging was performed on an Axiovert inverted fluorescence microscope (Carl Zeiss), equipped with a Yokogawa spinning disk, a CoolSNAP HQ2 14-bit camera (PhotoMetrics), and laser lines for 488 nm (40% laser power, 400-ms excitation) and 561 nm (40% laser power, 200-ms excitation). To facilitate time-lapse imaging, the microscope has a programmable stage with definite focus and a stage enclosure that maintains samples at 37 °C and 5% CO2 with humidity. Images were captured every 15 min. For each position a five-by-five X–Y grid was sampled with five z-positions at 2.5-μm intervals. The objective used was 40× oil, 1.3 N.A.
Cell Segmentation and Image Analysis.
For analyzing the MIEP dynamics after infection, a maximal intensity projection was applied to all collected z planes of the same field of view to increase signal intensity. Single-cell tracking and segmentation were performed with custom-written code in MATLAB (MathWorks) as described (40). mCherry and EYFP intensities were calculated as the median fluorescence intensity of each segmented cell. Single trajectories were smoothed using a qubic spline (MATLAB) and synchronized in silico to the first detection of fluorescence to account for variation in MIEP expression start time between cells. Cells that initiated expression more than 12 h after virus was removed or that tracked for less than 10 h were discarded from the analysis.
For quantification of pp71-EYFP degradation rate, the number of pp71-EYFP foci were counted at each time point using ImageJ software by the following steps. A maximal z-projection was applied to all z-stacks planes of the same field of view to increase signal strength, then a uniform threshold was set for all images, and the number of EYFP dots was counted. Cells were stained with DAPI to quantify the number of cells for each field of view.
qPCR.
ARPE-19 cells cultured were infected with IE1-CMDR virus at MOI = 0.1, cultured in medium with 1 μM Shield-1 or without Shield-1, and harvested 48 h postinfection. Total RNA was extracted using an RNEASY RNA Isolation Kit (74104; QIAGEN) and reverse-transcribed using a QuantiTet Reverse Transcription Kit (205311; QIAGEN). CDNA samples were diluted 1:40 and analyzed on a 7900HT Fast Real-Time PCR System (4329003; Thermo-Fisher) using designed primers (SI Appendix, Table S1) and Fast SYBR Green Master Mix (4385612; Applied Biosystems). Relative mRNA levels of mCherry and EYFP were quantified using peptidylprolyl isomerase A (PP1A) as a reference gene.
Flow Cytometry and Calculation of Degradation Rates.
To quantify IE1 and IE2 degradation rates ARPE-19 cells were seeded in a six-well plate, grown overnight, and then infected on ice with IE1-CMDR virus (MOI = 0.1) in the presence of 1 μM Shield-1. After infection cells were cultured in fresh medium supplemented with 1 μM Shield-1 for 24 h. After 24 h, cells were washed twice with fresh medium and cultured in medium containing 50 μg/mL cycloheximide (Sigma-Aldrich), to stop protein translation, either with 1 μM Shield-1 or without Shield-1. Cells were harvested at indicated time points and mCherry and EYFP signals measured on an LSRII flow cytometer. Infected cells were defined based on mCherry gating and mean fluorescence intensities of mCherry and EYFP were used to calculate degradation rates by fitting the data to an exponential decay curve (SI Appendix, Fig. S4).
Calculations of Autocorrelation Function.
HF-ACFs were calculated as described in ref. 40 to estimate the change in positive-feedback strength. Positive feedback is predicted to increase the correlation time of the noise HF-ACF by an amount related directly to the feedback strength (40). Derivation of ACFs was done as follows. First, for each cell m, fluorescence trajectories were detrended (normalized) by subtracting from each trajectory the population time-dependent average fluorescence (i.e., the general trend), to account for gene expression changes that affect all of the cells (i.e., high-frequency filtering to isolate intrinsic noise). Second, for a given cell m the HF-ACF was calculated from
where N denotes the normalized fluorescence trajectory and j has integer values from 0 to K−1. Next, for M cell trajectories the HF-ACF is calculated as
For a full derivation, we refer the reader to ref. 40.
Mathematical Modeling and Simulations.
The differential equations describing the dynamics of IE1 and pp71 in the main text were simulated with MATLAB (MathWorks) using the ODE45 solver. STI (sustained-to-transient index) was defined as the ratio between IE1 expression at 24 h postinfection divided by the maximal IE1 expression and examined over a range of parameters. See SI Appendix, Tables S2 and S3 for parameters values.
The stochastic model of IE1 and pp71 described in SI Appendix, Fig. S6 and Table S4 was simulated in MATLAB by implementing a stochastic Gillespie algorithm (51). For each parameter set, 500 trajectories were simulated over a virtual time course of 15 h.
Supplementary Material
Acknowledgments
We thank Thomas Stamminger for the generous contribution of the EYFP-pp71 reporter virus, Cynthia Bolovan-Fritts for support and assistance, Victoria Saykally for technical assistance, Marielle Cavrois for technical expertise, and members of our group for fruitful discussions. L.S.W. acknowledges support from the Bowes Distinguished Professorship, the Alfred P. Sloan Research Fellowship, NIH Director’s New Innovator Award OD006677 and Pioneer Award OD17181 programs, and Gladstone Flow Cytometry Core, funded through NIH P30 AI027763.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802905115/-/DCSupplemental.
References
- 1.Bardeen J, Brattain WH. The transistor, a semi-conductor triode. Phys Rev. 1948;74:230–231. [Google Scholar]
- 2.Becker JA, Shive JN. The transistor–A new semiconductor amplifier. Electr Eng. 1949;68:215–221. [Google Scholar]
- 3.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 4.Monod J, Jacob F. Teleonomic mechanisms in cellular metabolism, growth, and differentiation. Cold Spring Harb Symp Quant Biol. 1961;26:389–401. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 5.Ferrell JE., Jr Self-perpetuating states in signal transduction: Positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 6.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 7.Hasty J, McMillen D, Isaacs F, Collins JJ. Computational studies of gene regulatory networks: In numero molecular biology. Nat Rev Genet. 2001;2:268–279. doi: 10.1038/35066056. [DOI] [PubMed] [Google Scholar]
- 8.Orchard RC, et al. Identification of F-actin as the dynamic hub in a microbial-induced GTPase polarity circuit. Cell. 2012;148:803–815. doi: 10.1016/j.cell.2011.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao J, Pilko A, Wollman R. Distinct cellular states determine calcium signaling response. Mol Syst Biol. 2016;12:894. doi: 10.15252/msb.20167137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampaio KL, Cavignac Y, Stierhof YD, Sinzger C. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol. 2005;79:2754–2767. doi: 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann H, Sindre H, Stamminger T. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol. 2002;76:5769–5783. doi: 10.1128/JVI.76.11.5769-5783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishov AM, et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79:5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bresnahan WA, Hultman GE, Shenk T. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J Virol. 2000;74:10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukac DM, Manuppello JR, Alwine JC. Transcriptional activation by the human cytomegalovirus immediate-early proteins: Requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis MJ, et al. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White EA, Clark CL, Sanchez V, Spector DH. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J Virol. 2004;78:1817–1830. doi: 10.1128/JVI.78.4.1817-1830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castillo JP, Yurochko AD, Kowalik TF. Role of human cytomegalovirus immediate-early proteins in cell growth control. J Virol. 2000;74:8028–8037. doi: 10.1128/jvi.74.17.8028-8037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy EA, Streblow DN, Nelson JA, Stinski MF. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J Virol. 2000;74:7108–7118. doi: 10.1128/jvi.74.15.7108-7118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn JH, Brignole EJ, 3rd, Hayward GS. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor RT, Bresnahan WA. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J Virol. 2006;80:920–928. doi: 10.1128/JVI.80.2.920-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresnahan WA, Shenk TE. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci USA. 2000;97:14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantrell SR, Bresnahan WA. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J Virol. 2005;79:7792–7802. doi: 10.1128/JVI.79.12.7792-7802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saffert RT, Kalejta RF. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol. 2006;80:3863–3871. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 28.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine JH, Fontes ME, Dworkin J, Elowitz MB. Pulsed feedback defers cellular differentiation. PLoS Biol. 2012;10:e1001252. doi: 10.1371/journal.pbio.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacArthur BD, et al. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat Cell Biol. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahrends R, et al. Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science. 2014;344:1384–1389. doi: 10.1126/science.1252079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos SD, Wollman R, Meyer T, Ferrell JE., Jr Spatial positive feedback at the onset of mitosis. Cell. 2012;149:1500–1513. doi: 10.1016/j.cell.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng MW, et al. An endogenous accelerator for viral gene expression confers a fitness advantage. Cell. 2012;151:1569–1580. doi: 10.1016/j.cell.2012.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski ES, Kemble GW, Lyle JM, Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavalai N, Kraiger M, Kaiser N, Stamminger T. Insertion of an EYFP-pp71 (UL82) coding sequence into the human cytomegalovirus genome results in a recombinant virus with enhanced viral growth. J Virol. 2008;82:10543–10555. doi: 10.1128/JVI.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson GW, Kelly C, Sinclair JH, Rickards C. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J Gen Virol. 1998;79:1233–1245. doi: 10.1099/0022-1317-79-5-1233. [DOI] [PubMed] [Google Scholar]
- 38.Lee HR, et al. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004;78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevels M, Brune W, Shenk T. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J Virol. 2004;78:7803–7812. doi: 10.1128/JVI.78.14.7803-7812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- 41.Cox CD, McCollum JM, Allen MS, Dar RD, Simpson ML. Using noise to probe and characterize gene circuits. Proc Natl Acad Sci USA. 2008;105:10809–10814. doi: 10.1073/pnas.0804829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 43.Schilling EM, et al. The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J Virol. 2017;91:e02049-16. doi: 10.1128/JVI.02049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White EA, Del Rosario CJ, Sanders RL, Spector DH. The IE2 60-kilodalton and 40-kilodalton proteins are dispensable for human cytomegalovirus replication but are required for efficient delayed early and late gene expression and production of infectious virus. J Virol. 2007;81:2573–2583. doi: 10.1128/JVI.02454-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pai A, Weinberger LS. Fate-regulating circuits in viruses: From discovery to new therapy targets. Annu Rev Virol. 2017;4:469–490. doi: 10.1146/annurev-virology-110615-035606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sachsenberg N, et al. Turnover of CD4+ and CD8+ T lymphocytes in HIV-1 infection as measured by Ki-67 antigen. J Exp Med. 1998;187:1295–1303. doi: 10.1084/jem.187.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Razooky BS, Pai A, Aull K, Rouzine IM, Weinberger LS. A hardwired HIV latency program. Cell. 2015;160:990–1001. doi: 10.1016/j.cell.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukashchuk V, McFarlane S, Everett RD, Preston CM. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J Virol. 2008;82:12543–12554. doi: 10.1128/JVI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Everett RD, et al. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Gent M, et al. Epstein-Barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with toll-like receptor signaling. PLoS Pathog. 2014;10:e1003960. doi: 10.1371/journal.ppat.1003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.