Figure 7.
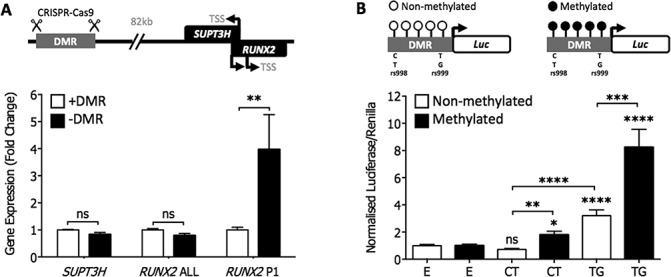
In vitro modulation of the SUPT3H/RUNX2-DMR. (A) CRISPR-Cas9 deletion of the DMR in the chondrocyte cell line Tc28a2 (TSS). Expression of SUPT3H, RUNX2 ALL (both isoforms) and RUNX2 P1 (long isoform) was measured by RT-qPCR. The graph represents the mean (+/− SEM) fold change of each gene/isoform following deletion of the DMR (−DMR) compared to control cells without deletion (+DMR). Each bar represents six biological repeat experiments. Statistical significance was determined by two-way ANOVA. **, P < 0.005; ns, not significant, P > 0.05. (B) Luciferase reporter analysis of DMR methylation in the chondrosarcoma cell line SW1353. Mean luciferase activity (21 technical repeats) of the DMR normalized against a control empty non-methylated or empty methylated vector. E is the empty vector control. CT denotes the non-risk conferring haplotype at rs62435998-rs62435999. TG denotes the risk-conferring haplotype at s62435998-rs62435999. Error bars represent the SEM. P-values were calculated using a Mann–Whitney two-tailed exact test and are compared to the relevant non-methylated or methylated empty vector control, unless otherwise indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
