Figure 1.
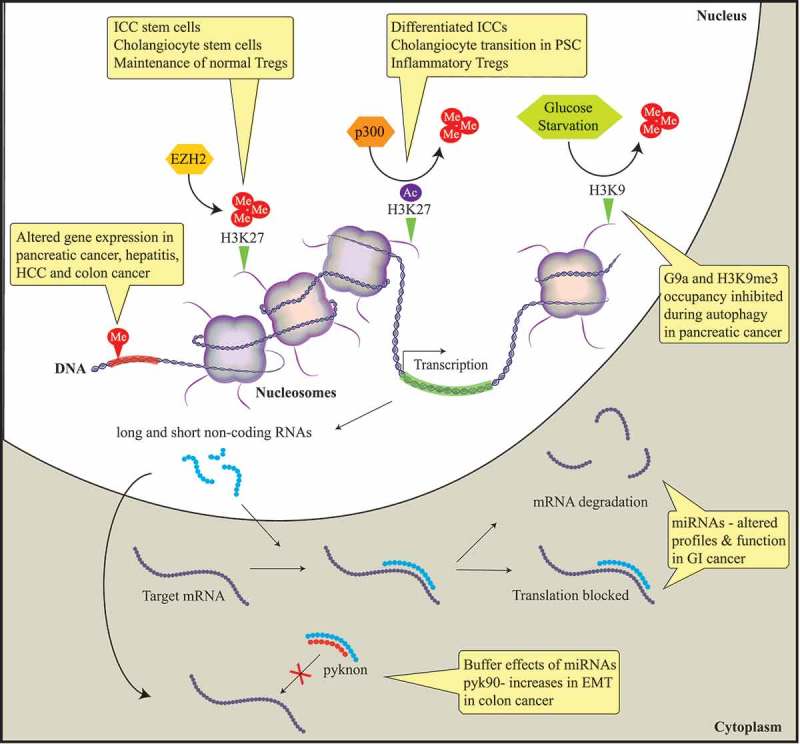
Diagram describing the main epigenetic mechanisms identified to date that have a role in the pathobiology of benign and malignant gastrointestinal diseases.
In this meeting report, we summarize and discuss presentations by international experts in the field of DNA methylation, histone-based epigenetics, and microRNAs. Epigenetic mechanisms that induce stable alterations to gene expression without alteration of the DNA sequence result in aberrant gene expression. Histone methylation alters gene expression in pancreatic cancer, hepatitis, hepatocellular cancer (HCC), and colon cancer. EZH2, a histone methyltransferase, induces the epigenetic gene silencing mark H3K27me3. This epigenetic mark mobilizes the progenitor interstitial cells of Cajal (ICC) to transition to mature ICC in gastrointestinal (GI) motility disorders. In addition, this mark maintains normal cholangiocytes and Regulatory T cells (T-regs) through FN1 and FOXP3 target silencing, respectively. The histone acetylase complex p300 replaces H3K27 methylation with acetylation resulting in FN1 transcription, promoting cholangiocytes to transition to primary sclerosing cholangitis (PSC) in liver disease. Autophagy in pancreatic cancer is associated with a depression of gene expression caused by the dissociation of the histone methyltransferase G9a and H3K9me3 from gene promoters. Non-protein-coding RNAs are associated with the initiation and progression of GI diseases through negatively regulating protein expression by binding to the promoter regions of targeted mRNAs leading to translational repression or mRNA degradation. Pyknons are lncRNAs that buffer the effects of miRNAs; pyk90 plays a role in epithelial-to-mesenchymal transition (EMT) in colon cancer via a targeted decreased expression of E-cadherin.
