Figure 5.
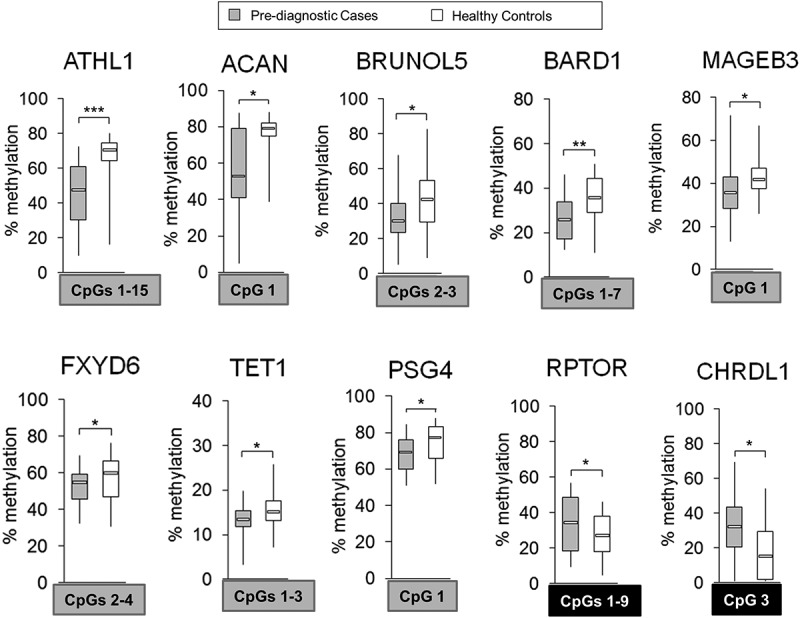
Differentially methylated regions within 10 probes corresponding to ATHL1, ACAN, BRUNOL5, BARD1, MAGEB3, FXYD6, TET1, PSG4, RPTOR, and CHRDL1 in pre-diagnostic hepatocellular carcinoma (HCC) cases vs. matched healthy controls. Average methylation level across selected CpG sites within a given probe in pre-diagnostic HCC cases (n = 21) vs. matched healthy controls (n = 21), as determined by pyrosequencing in 10 probes corresponding to 10 genes: ATHL1, ACAN, BRUNOL5, BARD1, MAGEB3, FXYD6, TET1, PSG4, RPTOR, and CHRDL1. Selected differentially methylated CpG sites were chosen based on showing consistent changes in DNA methylation in the tested region and are shown in square box, with grey indicating hypomethylation and black indicating hypermethylation in cases vs. controls. ***P <0.001, **P <0.01, *P <0.05.
