Figure 6.
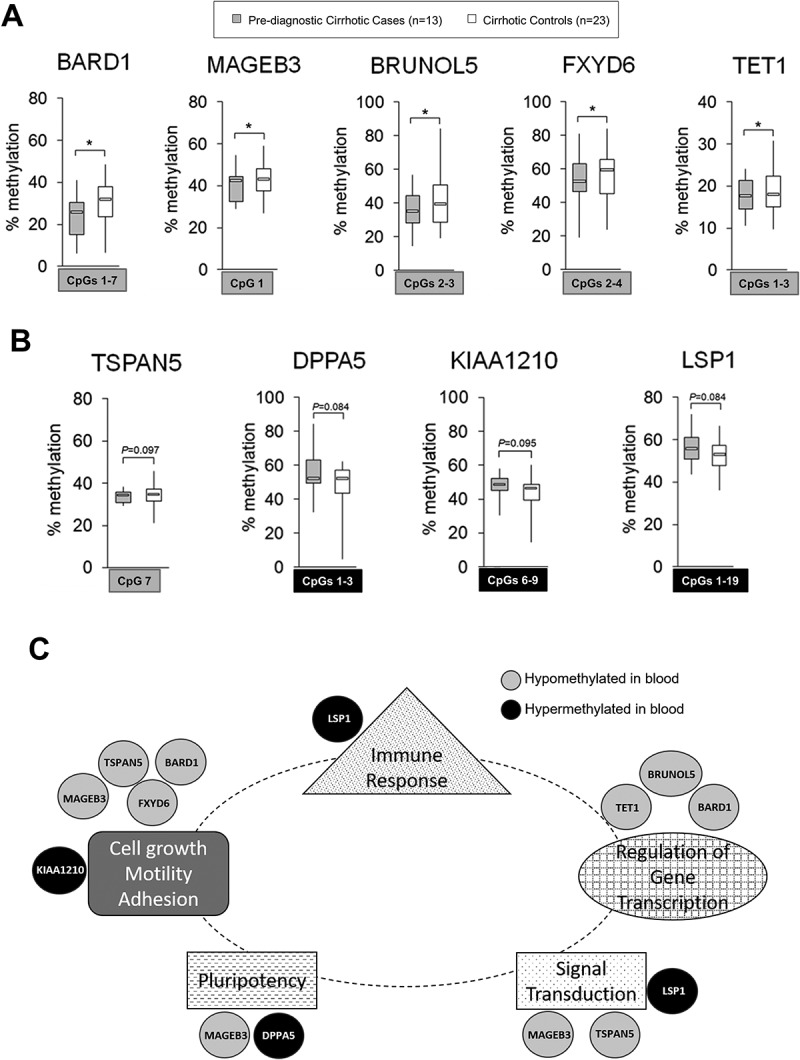
Differentially methylated regions within 9 probes: BARD1, MAGEB3, BRUNOL5, FXYD6, TET1, TSPAN5, DPPA5, KIAA1210, and LSP1, in pre-diagnostic hepatocellular carcinoma (HCC) cases with underlying liver cirrhosis vs. cirrhotic controls free of cancer: candidate probes for HCC early detection in populations at risk. (A, B) Average methylation level across selected CpG sites within: A) BARD1, MAGEB3, BRUNOL5, FXYD6, and TET1 in pre-diagnostic cirrhotic HCC cases (light grey, n = 13 from Study Group #3) and cirrhotic controls free of cancer (white, n = 23); B) TSPAN5, DPPA5, KIAA1210, and LSP1 in pre-diagnostic cirrhotic HCC cases (light grey, n = 13 from Study Group #3) and cirrhotic controls that remained cancer free within the same period of follow up (white, n = 23), as determined by pyrosequencing. Selected differentially methylated CpG sites are shown in square box, with grey indicating hypomethylation and black indicating hypermethylation in cases vs. controls. ***P <0.001, **P <0.01, *P <0.05. (C) A network map of genes corresponding to 9 probes that can distinguish cirrhotic patients who will develop hepatocellular carcinoma (HCC) from those who will stay cancer free within a given follow up period. The network was established based on publicly available data and shows the implication of the genes in functions related to cells actively proliferating and undergoing functional changes (e.g., immune cells or cancer cells). Genes hypomethylated in blood samples of pre-diagnostic hepatocellular carcinoma (HCC) cases such as BARD1, MAGEB3, BRUNOL5, FXYD6, TET1, and TSPAN5 (marked in grey) show evidence for the oncogenic role whereas hypermethylated genes including DPPA5, KIAA1210, and LSP1 (marked in black) are implicated in tumor suppressor functions. Several of them such as BARD1, FXYD6, TSPAN5, and LSP1 are functionally related to immune pathways as indicated by ImmuNet, a collection of immune-related functional relationships networks. TSPAN5 was also found to be expressed in T-lymphoid cells [46].
